Niche stiffness underlies the ageing of central nervous system progenitor cells
- PMID: 31413369
- PMCID: PMC7025879
- DOI: 10.1038/s41586-019-1484-9
Niche stiffness underlies the ageing of central nervous system progenitor cells
Erratum in
-
Author Correction: Niche stiffness underlies the ageing of central nervous system progenitor cells.Nature. 2019 Sep;573(7773):E3. doi: 10.1038/s41586-019-1552-1. Nature. 2019. PMID: 31462780
Abstract
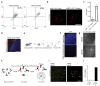
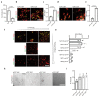
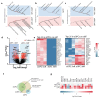
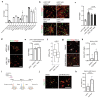
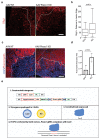
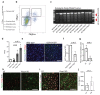
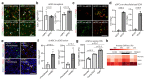
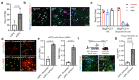
Ageing causes a decline in tissue regeneration owing to a loss of function of adult stem cell and progenitor cell populations1. One example is the deterioration of the regenerative capacity of the widespread and abundant population of central nervous system (CNS) multipotent stem cells known as oligodendrocyte progenitor cells (OPCs)2. A relatively overlooked potential source of this loss of function is the stem cell 'niche'-a set of cell-extrinsic cues that include chemical and mechanical signals3,4. Here we show that the OPC microenvironment stiffens with age, and that this mechanical change is sufficient to cause age-related loss of function of OPCs. Using biological and synthetic scaffolds to mimic the stiffness of young brains, we find that isolated aged OPCs cultured on these scaffolds are molecularly and functionally rejuvenated. When we disrupt mechanical signalling, the proliferation and differentiation rates of OPCs are increased. We identify the mechanoresponsive ion channel PIEZO1 as a key mediator of OPC mechanical signalling. Inhibiting PIEZO1 overrides mechanical signals in vivo and allows OPCs to maintain activity in the ageing CNS. We also show that PIEZO1 is important in regulating cell number during CNS development. Thus we show that tissue stiffness is a crucial regulator of ageing in OPCs, and provide insights into how the function of adult stem and progenitor cells changes with age. Our findings could be important not only for the development of regenerative therapies, but also for understanding the ageing process itself.
Conflict of interest statement
Figures

Similar articles
-
Sox2 Sustains Recruitment of Oligodendrocyte Progenitor Cells following CNS Demyelination and Primes Them for Differentiation during Remyelination.J Neurosci. 2015 Aug 19;35(33):11482-99. doi: 10.1523/JNEUROSCI.3655-14.2015. J Neurosci. 2015. PMID: 26290228 Free PMC article.
-
Mechanical environment modulates biological properties of oligodendrocyte progenitor cells.Stem Cells Dev. 2012 Nov 1;21(16):2905-14. doi: 10.1089/scd.2012.0189. Epub 2012 Jul 3. Stem Cells Dev. 2012. PMID: 22646081 Free PMC article.
-
Decline in rate of colonization of oligodendrocyte progenitor cell (OPC)-depleted tissue by adult OPCs with age.J Neuropathol Exp Neurol. 2003 Sep;62(9):908-16. doi: 10.1093/jnen/62.9.908. J Neuropathol Exp Neurol. 2003. PMID: 14533780
-
Engineering biomaterial microenvironments to promote myelination in the central nervous system.Brain Res Bull. 2019 Oct;152:159-174. doi: 10.1016/j.brainresbull.2019.07.013. Epub 2019 Jul 12. Brain Res Bull. 2019. PMID: 31306690 Review.
-
Mechanical forces direct stem cell behaviour in development and regeneration.Nat Rev Mol Cell Biol. 2017 Dec;18(12):728-742. doi: 10.1038/nrm.2017.108. Epub 2017 Nov 8. Nat Rev Mol Cell Biol. 2017. PMID: 29115301 Free PMC article. Review.
Cited by
-
Disruption of oligodendrocyte progenitor cells is an early sign of pathology in the triple transgenic mouse model of Alzheimer's disease.Neurobiol Aging. 2020 Oct;94:130-139. doi: 10.1016/j.neurobiolaging.2020.05.016. Epub 2020 Jun 7. Neurobiol Aging. 2020. PMID: 32619874 Free PMC article.
-
Mechanisms of mechanotransduction and physiological roles of PIEZO channels.Nat Rev Mol Cell Biol. 2024 Nov;25(11):886-903. doi: 10.1038/s41580-024-00773-5. Epub 2024 Sep 9. Nat Rev Mol Cell Biol. 2024. PMID: 39251883 Review.
-
Substrate stiffness induced mechanotransduction regulates temporal evolution of human fetal neural progenitor cell phenotype, differentiation, and biomechanics.Biomater Sci. 2020 Sep 30;8(19):5452-5464. doi: 10.1039/d0bm01349h. Biomater Sci. 2020. PMID: 32996962 Free PMC article.
-
Aging and Rejuvenation of Neural Stem Cells and Their Niches.Cell Stem Cell. 2020 Aug 6;27(2):202-223. doi: 10.1016/j.stem.2020.07.002. Epub 2020 Jul 28. Cell Stem Cell. 2020. PMID: 32726579 Free PMC article. Review.
-
Nutrition Interventions of Herbal Compounds on Cellular Senescence.Oxid Med Cell Longev. 2022 Apr 27;2022:1059257. doi: 10.1155/2022/1059257. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35528514 Free PMC article. Review.
References
-
- Goodell MA, Rando TA. Stem cells and healthy aging. Science. 2015;350:1199–1204. - PubMed
-
- Gopinath SD, Rando TA. Stem Cell Review Series: Aging of the skeletal muscle stem cell niche. Aging Cell. 2008;7:590–598. - PubMed
-
- Hinks GL, Franklin RJ. Delayed changes in growth factor gene expression during slow remyelination in the CNS of aged rats. Mol Cell Neurosci. 2000;16:542–556. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

