Residue-specific identification of phase separation hot spots of Alzheimer's-related protein tau
- PMID: 31341602
- PMCID: PMC6610569
- DOI: 10.1039/c9sc00531e
Residue-specific identification of phase separation hot spots of Alzheimer's-related protein tau
Abstract
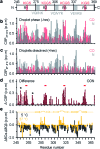
Liquid-liquid phase separation (LLPS) of proteins enables the formation of non-membrane-bound organelles in cells and is associated with cancer and neurodegeneration. Little is known however about the structure and dynamics of proteins in LLPS conditions, because of the polymorphic nature of liquid-like protein droplets. Using carbon-detected NMR experiments we here show that the conversion of the aggregation-prone repeat region of the Alzheimer's-related protein tau from the dispersed monomeric state to phase-separated liquid-like droplets involves tau's aggregation-prone hexapeptides and regulatory KXGS motifs. Droplet dissolution in presence of 1,6-hexanediol revealed that chemical shift perturbations in the hexapeptide motifs are temperature driven, while those in KXGS motifs report on phase separation. Residue-specific secondary structure analysis further indicated that tau's repeat region exists in extended conformation in the dispersed state and attains transient β-hairpin propensity upon LLPS. Taken together our work shows that NMR spectroscopy can provide high-resolution insights into LLPS-induced changes in intrinsically disordered proteins.
Figures
Similar articles
-
Liquid-Liquid Phase Separation of Tau Protein Is Encoded at the Monomeric Level.J Phys Chem Lett. 2021 Mar 18;12(10):2576-2586. doi: 10.1021/acs.jpclett.1c00208. Epub 2021 Mar 9. J Phys Chem Lett. 2021. PMID: 33686854
-
Liquid-liquid phase separation of tau protein: The crucial role of electrostatic interactions.J Biol Chem. 2019 Jul 19;294(29):11054-11059. doi: 10.1074/jbc.AC119.009198. Epub 2019 May 16. J Biol Chem. 2019. PMID: 31097543 Free PMC article.
-
EFhd2 Affects Tau Liquid-Liquid Phase Separation.Front Neurosci. 2019 Aug 13;13:845. doi: 10.3389/fnins.2019.00845. eCollection 2019. Front Neurosci. 2019. PMID: 31456657 Free PMC article.
-
Tau Condensates.Adv Exp Med Biol. 2019;1184:327-339. doi: 10.1007/978-981-32-9358-8_24. Adv Exp Med Biol. 2019. PMID: 32096047 Review.
-
Temperature, Hydrostatic Pressure, and Osmolyte Effects on Liquid-Liquid Phase Separation in Protein Condensates: Physical Chemistry and Biological Implications.Chemistry. 2019 Oct 11;25(57):13049-13069. doi: 10.1002/chem.201902210. Epub 2019 Aug 22. Chemistry. 2019. PMID: 31237369 Review.
Cited by
-
The structure and phase of tau: from monomer to amyloid filament.Cell Mol Life Sci. 2021 Mar;78(5):1873-1886. doi: 10.1007/s00018-020-03681-x. Epub 2020 Oct 19. Cell Mol Life Sci. 2021. PMID: 33078207 Free PMC article. Review.
-
1H, 13C and 15N resonance assignments of stress granule key component G3BP1 RRM domain.Biomol NMR Assign. 2022 Apr;16(1):109-111. doi: 10.1007/s12104-022-10067-6. Epub 2022 Feb 12. Biomol NMR Assign. 2022. PMID: 35150414 Free PMC article.
-
Conformations of a Low-Complexity Protein in Homogeneous and Phase-Separated Frozen Solutions.bioRxiv [Preprint]. 2024 Jul 25:2024.07.25.605144. doi: 10.1101/2024.07.25.605144. bioRxiv. 2024. Update in: Biophys J. 2024 Dec 3;123(23):4097-4114. doi: 10.1016/j.bpj.2024.11.001 PMID: 39372747 Free PMC article. Updated. Preprint.
-
Tau liquid-liquid phase separation: At the crossroads of tau physiology and tauopathy.J Cell Physiol. 2024 Jun;239(6):e30853. doi: 10.1002/jcp.30853. Epub 2022 Aug 18. J Cell Physiol. 2024. PMID: 35980344 Review.
-
Liquid-liquid phase separation induces pathogenic tau conformations in vitro.Nat Commun. 2020 Jun 4;11(1):2809. doi: 10.1038/s41467-020-16580-3. Nat Commun. 2020. PMID: 32499559 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

