Liquid biopsy-based comprehensive gene mutation profiling for gynecological cancer using CAncer Personalized Profiling by deep Sequencing
- PMID: 31320709
- PMCID: PMC6639322
- DOI: 10.1038/s41598-019-47030-w
Liquid biopsy-based comprehensive gene mutation profiling for gynecological cancer using CAncer Personalized Profiling by deep Sequencing
Abstract
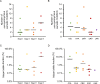
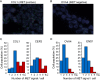
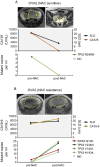
Liquid biopsies of circulating tumor DNA (ctDNA) have recently been used as a non-invasive diagnostic tool for detecting tumor-specific mutations. We present a study of ctDNA liquid biopsies in gynecological cancer using an ultrasensitive next-generation sequencing-based method for ctDNA detection named CAncer Personalized Profiling by deep Sequencing (CAPP-Seq). We performed CAPP-Seq with plasma-ctDNA obtained from 16 patients with gynecological cancer. In all cases, at least one non-synonymous somatic mutation was detected in the ctDNA. In the pre-treatment ctDNA, 4 of 16, 4/16, 5/16, 2/16, 2/16, and 2/16 patients had TP53, KRAS, APC, PIK3CA, BRCA1, and EGFR mutations, respectively. MET gene copy-number gains were detected in the ctDNA of 2 of 16 patients, and FISH analysis of the paired tumor samples confirmed these results. In 2 neoadjuvant chemotherapy-treated ovarian cancer patients, the changes in gene mutation patterns were associated with the treatment response. These findings suggest that CAPP-Seq-based liquid biopsies can be used for the genetic characterization of independent gynecological cancers with high frequency, and might be clinically useful for non-invasive tumor genotyping and therapeutic response monitoring.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Circulating Tumor DNA (ctDNA) and Its Role in Gynecologic Malignancies.Curr Treat Options Oncol. 2024 Apr;25(4):510-522. doi: 10.1007/s11864-024-01180-w. Epub 2024 Mar 12. Curr Treat Options Oncol. 2024. PMID: 38472567 Review.
-
Changes in the gene mutation profiles of circulating tumor DNA detected using CAPP-Seq in neoadjuvant chemotherapy-treated advanced ovarian cancer.Oncol Lett. 2020 Apr;19(4):2713-2720. doi: 10.3892/ol.2020.11356. Epub 2020 Jan 28. Oncol Lett. 2020. PMID: 32218822 Free PMC article.
-
Detection of Circulating Tumor DNA in Lymphoma Patients.Methods Mol Biol. 2025;2865:475-490. doi: 10.1007/978-1-0716-4188-0_21. Methods Mol Biol. 2025. PMID: 39424738
-
Monitoring of cancer patients via next-generation sequencing of patient-derived circulating tumor cells and tumor DNA.Cancer Sci. 2019 Aug;110(8):2590-2599. doi: 10.1111/cas.14092. Epub 2019 Jul 23. Cancer Sci. 2019. PMID: 31169336 Free PMC article.
-
Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq.Expert Rev Mol Diagn. 2015 Jun;15(6):715-9. doi: 10.1586/14737159.2015.1019476. Epub 2015 Mar 16. Expert Rev Mol Diagn. 2015. PMID: 25773944 Free PMC article. Review.
Cited by
-
Liquid Biopsy in Colorectal Carcinoma: Clinical Applications and Challenges.Cancers (Basel). 2020 May 27;12(6):1376. doi: 10.3390/cancers12061376. Cancers (Basel). 2020. PMID: 32471160 Free PMC article. Review.
-
Personalized tumor-specific DNA junctions to detect circulating tumor in patients with endometrial cancer.PLoS One. 2021 Jun 10;16(6):e0252390. doi: 10.1371/journal.pone.0252390. eCollection 2021. PLoS One. 2021. PMID: 34111149 Free PMC article.
-
Clinical implications of plasma circulating tumor DNA in gynecologic cancer patients.Mol Oncol. 2021 Jan;15(1):67-79. doi: 10.1002/1878-0261.12791. Epub 2020 Sep 17. Mol Oncol. 2021. PMID: 32881280 Free PMC article.
-
Molecular Guided Treatments in Gynecologic Oncology: Analysis of a Real-World Precision Cancer Medicine Platform.Oncologist. 2020 Jul;25(7):e1060-e1069. doi: 10.1634/theoncologist.2019-0904. Epub 2020 May 8. Oncologist. 2020. PMID: 32369643 Free PMC article.
-
Predicting osimertinib-treatment outcomes through EGFR mutant-fraction monitoring in the circulating tumor DNA of EGFR T790M-positive patients with non-small cell lung cancer (WJOG8815L).Mol Oncol. 2021 Jan;15(1):126-137. doi: 10.1002/1878-0261.12841. Epub 2020 Nov 17. Mol Oncol. 2021. PMID: 33131198 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

