The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells
- PMID: 31229354
- PMCID: PMC6642154
- DOI: 10.1016/j.immuni.2019.05.004
The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells
Abstract
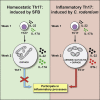
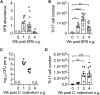
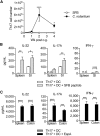
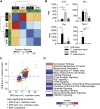
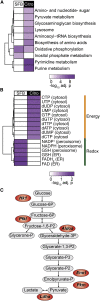
T helper 17 (Th17) cells are pathogenic in many inflammatory diseases, but also support the integrity of the intestinal barrier in a non-inflammatory manner. It is unclear what distinguishes inflammatory Th17 cells elicited by pathogens and tissue-resident homeostatic Th17 cells elicited by commensals. Here, we compared the characteristics of Th17 cells differentiating in response to commensal bacteria (SFB) to those differentiating in response to a pathogen (Citrobacter rodentium). Homeostatic Th17 cells exhibited little plasticity towards expression of inflammatory cytokines, were characterized by a metabolism typical of quiescent or memory T cells, and did not participate in inflammatory processes. In contrast, infection-induced Th17 cells showed extensive plasticity towards pro-inflammatory cytokines, disseminated widely into the periphery, and engaged aerobic glycolysis in addition to oxidative phosphorylation typical for inflammatory effector cells. These findings will help ensure that future therapies directed against inflammatory Th17 cells do not inadvertently damage the resident gut population.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Polarizing the T helper 17 response in Citrobacter rodentium infection via expression of resistin-like molecule α.Gut Microbes. 2014 May-Jun;5(3):363-8. doi: 10.4161/gmic.29100. Epub 2014 May 15. Gut Microbes. 2014. PMID: 24831469 Free PMC article.
-
Citrobacter rodentium Induces Tissue-Resident Memory CD4+ T Cells.Infect Immun. 2019 Jun 20;87(7):e00295-19. doi: 10.1128/IAI.00295-19. Print 2019 Jul. Infect Immun. 2019. PMID: 31061145 Free PMC article.
-
Regulatory T cells promote a protective Th17-associated immune response to intestinal bacterial infection with C. rodentium.Mucosal Immunol. 2014 Nov;7(6):1290-301. doi: 10.1038/mi.2014.17. Epub 2014 Mar 19. Mucosal Immunol. 2014. PMID: 24646939
-
The dichotomous nature of T helper 17 cells.Nat Rev Immunol. 2017 Sep;17(9):535-544. doi: 10.1038/nri.2017.50. Epub 2017 May 30. Nat Rev Immunol. 2017. PMID: 28555673 Review.
-
Th17 plasticity and its relevance to inflammatory bowel disease.J Autoimmun. 2018 Feb;87:38-49. doi: 10.1016/j.jaut.2017.12.004. Epub 2017 Dec 28. J Autoimmun. 2018. PMID: 29290521 Review.
Cited by
-
MicroRNA-221 and -222 modulate intestinal inflammatory Th17 cell response as negative feedback regulators downstream of interleukin-23.Immunity. 2021 Mar 9;54(3):514-525.e6. doi: 10.1016/j.immuni.2021.02.015. Epub 2021 Mar 2. Immunity. 2021. PMID: 33657395 Free PMC article.
-
Th17 Cells, Glucocorticoid Resistance, and Depression.Cells. 2023 Nov 30;12(23):2749. doi: 10.3390/cells12232749. Cells. 2023. PMID: 38067176 Free PMC article. Review.
-
Dietary cellulose induces anti-inflammatory immunity and transcriptional programs via maturation of the intestinal microbiota.Gut Microbes. 2020 Nov 9;12(1):1-17. doi: 10.1080/19490976.2020.1829962. Gut Microbes. 2020. PMID: 33079623 Free PMC article.
-
Mucosal Immunity and HIV Acquisition in Women.Curr Opin Physiol. 2021 Feb;19:32-38. doi: 10.1016/j.cophys.2020.07.021. Epub 2020 Aug 18. Curr Opin Physiol. 2021. PMID: 33103019 Free PMC article.
-
M1 macrophages may be effective adjuvants for promoting Th‑17 differentiation in HBeAg positive hepatitis patients with ALT ≤2ULN.Mol Med Rep. 2023 Mar;27(3):63. doi: 10.3892/mmr.2023.12950. Epub 2023 Feb 3. Mol Med Rep. 2023. PMID: 36734259 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

