HIV-1 DNA sequence diversity and evolution during acute subtype C infection
- PMID: 31227699
- PMCID: PMC6588551
- DOI: 10.1038/s41467-019-10659-2
HIV-1 DNA sequence diversity and evolution during acute subtype C infection
Abstract
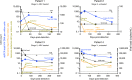
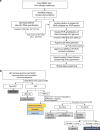
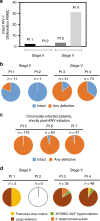
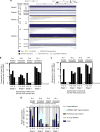
Little is known about the genotypic make-up of HIV-1 DNA genomes during the earliest stages of HIV-1 infection. Here, we use near-full-length, single genome next-generation sequencing to longitudinally genotype and quantify subtype C HIV-1 DNA in four women identified during acute HIV-1 infection in Durban, South Africa, through twice-weekly screening of high-risk participants. In contrast to chronically HIV-1-infected patients, we found that at the earliest phases of infection in these four participants, the majority of viral DNA genomes are intact, lack APOBEC-3G/F-associated hypermutations, have limited genome truncations, and over one year show little indication of cytotoxic T cell-driven immune selections. Viral sequence divergence during acute infection is predominantly fueled by single-base substitutions and is limited by treatment initiation during the earliest stages of disease. Our observations provide rare longitudinal insights of HIV-1 DNA sequence profiles during the first year of infection to inform future HIV cure research.
Conflict of interest statement
M.L. has received speaking and consulting honoraria from Merck & Co., Inc. and Gilead Sciences Inc. T.N. receives research funding support from Gilead Sciences Inc., a pharmaceutical company with interests in HIV cure research. The other authors declare no competing interests.
Figures
Similar articles
-
Rapid, complex adaptation of transmitted HIV-1 full-length genomes in subtype C-infected individuals with differing disease progression.AIDS. 2013 Feb 20;27(4):507-18. doi: 10.1097/QAD.0b013e32835cab64. AIDS. 2013. PMID: 23370465 Free PMC article.
-
Evidence of susceptibility to lamivudine-based HAART and genetic stability of hepatitis B virus (HBV) in HIV co-infected patients: A South African longitudinal HBV whole genome study.Infect Genet Evol. 2016 Sep;43:232-8. doi: 10.1016/j.meegid.2016.05.035. Epub 2016 May 28. Infect Genet Evol. 2016. PMID: 27245151
-
Analysis of drug resistance mutations in whole blood DNA from HIV-1 infected patients by single genome and ultradeep sequencing analysis.J Virol Methods. 2018 Oct;260:1-5. doi: 10.1016/j.jviromet.2018.06.020. Epub 2018 Jun 30. J Virol Methods. 2018. PMID: 29969601
-
The challenge of HIV-1 subtype diversity.N Engl J Med. 2008 Apr 10;358(15):1590-602. doi: 10.1056/NEJMra0706737. N Engl J Med. 2008. PMID: 18403767 Free PMC article. Review. No abstract available.
-
Impact of HIV-1 pol diversity on drug resistance and its clinical implications.Curr Opin Infect Dis. 2006 Dec;19(6):594-606. doi: 10.1097/QCO.0b013e3280109122. Curr Opin Infect Dis. 2006. PMID: 17075337 Review.
Cited by
-
Near-Full-Length Single-Genome HIV-1 DNA Sequencing.Methods Mol Biol. 2022;2407:357-364. doi: 10.1007/978-1-0716-1871-4_23. Methods Mol Biol. 2022. PMID: 34985675 Free PMC article.
-
Next-Generation Sequencing in a Direct Model of HIV Infection Reveals Important Parallels to and Differences from In Vivo Reservoir Dynamics.J Virol. 2020 Apr 16;94(9):e01900-19. doi: 10.1128/JVI.01900-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32051279 Free PMC article.
-
HIV-1 reservoir evolution in infants infected with clade C from Mozambique.Int J Infect Dis. 2023 Feb;127:129-136. doi: 10.1016/j.ijid.2022.11.042. Epub 2022 Dec 5. Int J Infect Dis. 2023. PMID: 36476348 Free PMC article.
-
Why and where an HIV cure is needed and how it might be achieved.Nature. 2019 Dec;576(7787):397-405. doi: 10.1038/s41586-019-1841-8. Epub 2019 Dec 18. Nature. 2019. PMID: 31853080 Free PMC article. Review.
-
Longitudinal Dynamics of Intact HIV Proviral DNA and Outgrowth Virus Frequencies in a Cohort of Individuals Receiving Antiretroviral Therapy.J Infect Dis. 2021 Jul 2;224(1):92-100. doi: 10.1093/infdis/jiaa718. J Infect Dis. 2021. PMID: 33216132 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

