Sodium sulfide selectively induces oxidative stress, DNA damage, and mitochondrial dysfunction and radiosensitizes glioblastoma (GBM) cells
- PMID: 31176262
- PMCID: PMC6556549
- DOI: 10.1016/j.redox.2019.101220
Sodium sulfide selectively induces oxidative stress, DNA damage, and mitochondrial dysfunction and radiosensitizes glioblastoma (GBM) cells
Abstract
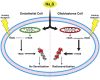
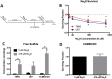
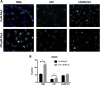
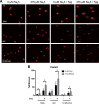
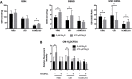
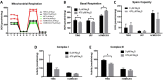
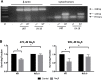
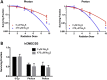
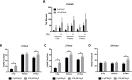
Glioblastoma (GBM) has a poor prognosis despite intensive treatment with surgery and chemoradiotherapy. Previous studies using dose-escalated radiotherapy have demonstrated improved survival; however, increased rates of radionecrosis have limited its use. Development of radiosensitizers could improve patient outcome. In the present study, we report the use of sodium sulfide (Na2S), a hydrogen sulfide (H2S) donor, to selectively kill GBM cells (T98G and U87) while sparing normal human cerebral microvascular endothelial cells (hCMEC/D3). Na2S also decreased mitochondrial respiration, increased oxidative stress and induced γH2AX foci and oxidative base damage in GBM cells. Since Na2S did not significantly alter T98G capacity to perform non-homologous end-joining or base excision repair, it is possible that GBM cell killing could be attributed to increased damage induction due to enhanced reactive oxygen species production. Interestingly, Na2S enhanced mitochondrial respiration, produced a more reducing environment and did not induce high levels of DNA damage in hCMEC/D3. Taken together, this data suggests involvement of mitochondrial respiration in Na2S toxicity in GBM cells. The fact that survival of LN-18 GBM cells lacking mitochondrial DNA (ρ0) was not altered by Na2S whereas the survival of LN-18 ρ+ cells was compromised supports this conclusion. When cells were treated with Na2S and photon or proton radiation, GBM cell killing was enhanced, which opens the possibility of H2S being a radiosensitizer. Therefore, this study provides the first evidence that H2S donors could be used in GBM therapy to potentiate radiation-induced killing.
Keywords: DNA damage; DNA repair; Glioblastoma; Hydrogen sulfide; Ionizing radiation; Mitochondria; Reactive oxygen species.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
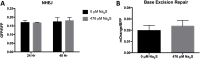
Similar articles
-
Radiosensitisation by pharmacological ascorbate in glioblastoma multiforme cells, human glial cells, and HUVECs depends on their antioxidant and DNA repair capabilities and is not cancer specific.Free Radic Biol Med. 2014 Sep;74:200-9. doi: 10.1016/j.freeradbiomed.2014.06.022. Epub 2014 Jun 30. Free Radic Biol Med. 2014. PMID: 24992837
-
Sirtuin 6 is essential for sodium sulfide-mediated cytoprotective effect in ischemia/reperfusion-stimulated brain endothelial cells.J Stroke Cerebrovasc Dis. 2015 Mar;24(3):601-9. doi: 10.1016/j.jstrokecerebrovasdis.2014.10.006. Epub 2014 Dec 24. J Stroke Cerebrovasc Dis. 2015. PMID: 25543188
-
Hydrogen sulfide attenuates homocysteine-induced neurotoxicity by preventing mitochondrial dysfunctions and oxidative damage: In vitro and in vivo studies.Neurochem Int. 2018 Nov;120:87-98. doi: 10.1016/j.neuint.2018.07.010. Epub 2018 Jul 26. Neurochem Int. 2018. PMID: 30055195
-
[Pathways for maintenance of mitochondrial DNA integrity and mitochondrial functions in cells exposed to ionizing radiation].Radiats Biol Radioecol. 2013 Mar-Apr;53(2):117-36. doi: 10.7868/s0869803113020045. Radiats Biol Radioecol. 2013. PMID: 23786028 Review. Russian.
-
Prodrugs of hydrogen sulfide and related sulfur species: recent development.Chin J Nat Med. 2020 Apr;18(4):296-307. doi: 10.1016/S1875-5364(20)30037-6. Chin J Nat Med. 2020. PMID: 32402407 Review.
Cited by
-
Leucine restriction ameliorates Fusobacterium nucleatum-driven malignant progression and radioresistance in nasopharyngeal carcinoma.Cell Rep Med. 2024 Oct 15;5(10):101753. doi: 10.1016/j.xcrm.2024.101753. Epub 2024 Oct 1. Cell Rep Med. 2024. PMID: 39357525 Free PMC article.
-
Inhibition of nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme of the nicotinamide adenine dinucleotide (NAD) salvage pathway, to target glioma heterogeneity through mitochondrial oxidative stress.Neuro Oncol. 2022 Feb 1;24(2):229-244. doi: 10.1093/neuonc/noab175. Neuro Oncol. 2022. PMID: 34260721 Free PMC article.
-
Gas Plasma Exposure of Glioblastoma Is Cytotoxic and Immunomodulatory in Patient-Derived GBM Tissue.Cancers (Basel). 2022 Feb 5;14(3):813. doi: 10.3390/cancers14030813. Cancers (Basel). 2022. PMID: 35159079 Free PMC article.
-
Altered Elemental Distribution in Male Rat Brain Tissue as a Predictor of Glioblastoma Multiforme Growth-Studies Using SR-XRF Microscopy.Int J Mol Sci. 2022 Jan 9;23(2):703. doi: 10.3390/ijms23020703. Int J Mol Sci. 2022. PMID: 35054889 Free PMC article.
-
Large-scale preparation of fluorescence multiplex host cell reactivation (FM-HCR) reporters.Nat Protoc. 2021 Sep;16(9):4265-4298. doi: 10.1038/s41596-021-00577-3. Epub 2021 Aug 6. Nat Protoc. 2021. PMID: 34363069 Free PMC article.
References
-
- Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. - PubMed
-
- Fitzek M.M., Thornton A.F., Rabinov J.D., Lev M.H., Pardo F.S., Munzenrider J.E. Accelerated fractionated proton/photon irradiation to 90 cobalt gray equivalent for glioblastoma multiforme: results of a phase II prospective trial. J. Neurosurg. 1999;91(2):251–260. - PubMed
-
- Iuchi T., Hatano K., Kodama T., Sakaida T., Yokoi S., Kawasaki K. Phase 2 trial of hypofractionated high-dose intensity modulated radiation therapy with concurrent and adjuvant temozolomide for newly diagnosed glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 2014;88(4):793–800. - PubMed
-
- Tanaka M., Ino Y., Nakagawa K., Tago M., Todo T. High-dose conformal radiotherapy for supratentorial malignant glioma: a historical comparison. Lancet Oncol. 2005;6(12):953–960. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

