Centrosomal Actin Assembly Is Required for Proper Mitotic Spindle Formation and Chromosome Congression
- PMID: 31096079
- PMCID: PMC6520610
- DOI: 10.1016/j.isci.2019.04.022
Centrosomal Actin Assembly Is Required for Proper Mitotic Spindle Formation and Chromosome Congression
Abstract
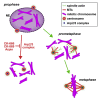
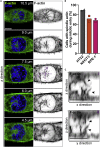
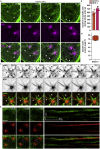
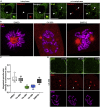
Cytoskeletal cross talk between actin filaments and microtubules is a common mechanism governing the assembly of cellular structures, i.e., during filopodia formation or cilia organization. However, potential actin-microtubule interactions during mammalian cell divisions are less well understood. At mitotic entry, centrosomes propagate the formation of the mitotic spindle, thereby aligning individual chromosomes to the metaphase plate, a process coined chromosome congression. Here, we identify actin filament assembly spatially defined at centrosomes contemporaneously with spindle microtubules forming during prometaphase. We show that pharmacological Arp2/3 complex inhibition as well as overexpression of the Arp2/3 complex inhibitory protein Arpin decreased spindle actin. As a consequence, mitotic spindle formation is impaired, which resulted in disorganized chromosome congression and ultimately mitotic defects in non-transformed cells. Thus centrosomal Arp2/3 complex activity plays a role in the maintenance of genomic integrity during mitosis.
Keywords: Biological Sciences; Cell Biology; Functional Aspects of Cell Biology; Organizational Aspects of Cell Biology.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Mechanisms of Chromosome Congression during Mitosis.Biology (Basel). 2017 Feb 17;6(1):13. doi: 10.3390/biology6010013. Biology (Basel). 2017. PMID: 28218637 Free PMC article. Review.
-
Prometaphase.Semin Cell Dev Biol. 2021 Sep;117:52-61. doi: 10.1016/j.semcdb.2021.06.004. Epub 2021 Jun 12. Semin Cell Dev Biol. 2021. PMID: 34127384 Review.
-
Cytochalasin J affects chromosome congression and spindle microtubule organization in PtK1 cells.Cell Motil Cytoskeleton. 1995;32(4):245-57. doi: 10.1002/cm.970320402. Cell Motil Cytoskeleton. 1995. PMID: 8608604
-
Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis.J Cell Sci. 1994 Apr;107 ( Pt 4):859-67. doi: 10.1242/jcs.107.4.859. J Cell Sci. 1994. PMID: 8056842
-
The microtubule-associated protein HURP recruits the centrosomal protein TACC3 to regulate K-fiber formation and support chromosome congression.J Biol Chem. 2018 Oct 5;293(40):15733-15747. doi: 10.1074/jbc.RA118.003676. Epub 2018 Jul 27. J Biol Chem. 2018. PMID: 30054275 Free PMC article.
Cited by
-
γ-Tubulin in microtubule nucleation and beyond.Front Cell Dev Biol. 2022 Sep 1;10:880761. doi: 10.3389/fcell.2022.880761. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36158181 Free PMC article. Review.
-
Genomic instability caused by Arp2/3 complex inactivation results in micronucleus biogenesis and cellular senescence.PLoS Genet. 2023 Jan 27;19(1):e1010045. doi: 10.1371/journal.pgen.1010045. eCollection 2023 Jan. PLoS Genet. 2023. PMID: 36706133 Free PMC article.
-
Spindle-F-actin interactions in mitotic spindles in an intact vertebrate epithelium.Mol Biol Cell. 2019 Jul 1;30(14):1645-1654. doi: 10.1091/mbc.E19-02-0126. Epub 2019 May 15. Mol Biol Cell. 2019. PMID: 31091161 Free PMC article.
-
Induced Arp2/3 Complex Depletion Increases FMNL2/3 Formin Expression and Filopodia Formation.Front Cell Dev Biol. 2021 Feb 1;9:634708. doi: 10.3389/fcell.2021.634708. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33598464 Free PMC article.
-
The regulation of actin dynamics during cell division and malignancy.Am J Cancer Res. 2021 Sep 15;11(9):4050-4069. eCollection 2021. Am J Cancer Res. 2021. PMID: 34659876 Free PMC article. Review.
References
-
- Baarlink C., Wang H., Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. - PubMed
- Baarlink, C., Wang, H. and Grosse, R. (2013) Nuclear actin network assembly by formins regulates the SRF coactivator MAL, Science, 340(), pp. 864-867. - PubMed
-
- Baarlink C., Plessner M., Sherrard A., Morita K., Misu S., Virant D., Kleinschnitz E.M., Harniman R., Alibhai D., Baumeister S. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 2017;19:1389–1399. - PubMed
- Baarlink, C., Plessner, M., Sherrard, A., Morita, K., Misu, S., Virant, D., Kleinschnitz, E. M., Harniman, R., Alibhai, D., Baumeister, S., Miyamoto, K., Endesfelder, U., Kaidi, A. and Grosse, R. (2017) A transient pool of nuclear F-actin at mitotic exit controls chromatin organization, Nat. Cell Biol., 19(), pp. 1389-1399. - PubMed
-
- Belin B.J., Lee T., Mullins R.D. DNA damage induces nuclear actin filament assembly by formin-2 and spire-1/2 that promotes efficient DNA repair. Elife. 2015;4:e07735. - PMC - PubMed
- Belin, B. J., Lee, T. and Mullins, R. D. (2015) DNA damage induces nuclear actin filament assembly by formin-2 and spire-1/2 that promotes efficient DNA repair, Elife, 4, e07735. - PMC - PubMed
-
- Caridi C.P., Dagostino C., Ryu T., Zapotoczny G., Delabaere L., Li X., Khodaverdian V.Y., Amaral N., Lin E., Rau A.R., Chiolo I. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature. 2018;559:54–60. - PMC - PubMed
- Caridi, C. P., Dagostino, C., Ryu, T., Zapotoczny, G., Delabaere, L., Li, X., Khodaverdian, V. Y., Amaral, N., Lin, E., Rau, A. R. and Chiolo, I. (2018) Nuclear F-actin and myosins drive relocalization of heterochromatic breaks, Nature, 559(), pp. 54-60. - PMC - PubMed
-
- Castagnetti S., Oliferenko S., Nurse P. Fission yeast cells undergo nuclear division in the absence of spindle microtubules. PLoS Biol. 2010;8:e1000512. - PMC - PubMed
- Castagnetti, S., Oliferenko, S. and Nurse, P. (2010) Fission yeast cells undergo nuclear division in the absence of spindle microtubules, PLoS Biol. Edited by M. Lichten. Public Library of Science, 8, p. e1000512 - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

