Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts
- PMID: 31043742
- PMCID: PMC6690743
- DOI: 10.1038/s41586-019-1173-8
Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts
Abstract
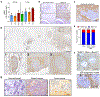
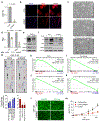
High-grade serous carcinoma has a poor prognosis, owing primarily to its early dissemination throughout the abdominal cavity. Genomic and proteomic approaches have provided snapshots of the proteogenomics of ovarian cancer1,2, but a systematic examination of both the tumour and stromal compartments is critical in understanding ovarian cancer metastasis. Here we develop a label-free proteomic workflow to analyse as few as 5,000 formalin-fixed, paraffin-embedded cells microdissected from each compartment. The tumour proteome was stable during progression from in situ lesions to metastatic disease; however, the metastasis-associated stroma was characterized by a highly conserved proteomic signature, prominently including the methyltransferase nicotinamide N-methyltransferase (NNMT) and several of the proteins that it regulates. Stromal NNMT expression was necessary and sufficient for functional aspects of the cancer-associated fibroblast (CAF) phenotype, including the expression of CAF markers and the secretion of cytokines and oncogenic extracellular matrix. Stromal NNMT expression supported ovarian cancer migration, proliferation and in vivo growth and metastasis. Expression of NNMT in CAFs led to depletion of S-adenosyl methionine and reduction in histone methylation associated with widespread gene expression changes in the tumour stroma. This work supports the use of ultra-low-input proteomics to identify candidate drivers of disease phenotypes. NNMT is a central, metabolic regulator of CAF differentiation and cancer progression in the stroma that may be therapeutically targeted.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


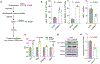



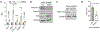



Similar articles
-
Nicotinamide N-methyltransferase overexpression may be associated with poor prognosis in ovarian cancer.J Obstet Gynaecol. 2021 Feb;41(2):248-253. doi: 10.1080/01443615.2020.1732891. Epub 2020 Apr 14. J Obstet Gynaecol. 2021. PMID: 32285726
-
High stromal nicotinamide N-methyltransferase (NNMT) indicates poor prognosis in colorectal cancer.Cancer Med. 2020 Mar;9(6):2030-2038. doi: 10.1002/cam4.2890. Epub 2020 Jan 27. Cancer Med. 2020. PMID: 31989785 Free PMC article.
-
Nicotinamide N-Methyltransferase Suppression Participates in Nickel-Induced Histone H3 Lysine9 Dimethylation in BEAS-2B Cells.Cell Physiol Biochem. 2017;41(5):2016-2026. doi: 10.1159/000475432. Epub 2017 Apr 13. Cell Physiol Biochem. 2017. PMID: 28420001
-
Nicotinamide N-methyltransferase: At the crossroads between cellular metabolism and epigenetic regulation.Mol Metab. 2021 Mar;45:101165. doi: 10.1016/j.molmet.2021.101165. Epub 2021 Jan 14. Mol Metab. 2021. PMID: 33453420 Free PMC article. Review.
-
Nicotinamide N-methyl transferase (NNMT): An emerging therapeutic target.Drug Discov Today. 2021 Nov;26(11):2699-2706. doi: 10.1016/j.drudis.2021.05.011. Epub 2021 May 21. Drug Discov Today. 2021. PMID: 34029690 Review.
Cited by
-
Nicotinamide N-Methyltransferase Remodeled Cell Metabolism and Aggravated Proinflammatory Responses by Activating STAT3/IL1β/PGE2 Pathway.ACS Omega. 2022 Oct 12;7(42):37509-37519. doi: 10.1021/acsomega.2c04286. eCollection 2022 Oct 25. ACS Omega. 2022. PMID: 36312432 Free PMC article.
-
Considerations of biomarker application for cancer continuum in the era of precision medicine.Curr Epidemiol Rep. 2022 Sep;9(3):200-211. doi: 10.1007/s40471-022-00295-8. Epub 2022 Jul 9. Curr Epidemiol Rep. 2022. PMID: 36090700 Free PMC article.
-
Immunobiology of high-grade serous ovarian cancer: lessons for clinical translation.Nat Rev Cancer. 2022 Nov;22(11):640-656. doi: 10.1038/s41568-022-00503-z. Epub 2022 Sep 15. Nat Rev Cancer. 2022. PMID: 36109621 Review.
-
Identification of Biological Functions and Prognostic Value of NNMT in Oral Squamous Cell Carcinoma.Biomolecules. 2022 Oct 15;12(10):1487. doi: 10.3390/biom12101487. Biomolecules. 2022. PMID: 36291696 Free PMC article.
-
The NADase enzyme CD38: an emerging pharmacological target for systemic sclerosis, systemic lupus erythematosus and rheumatoid arthritis.Curr Opin Rheumatol. 2020 Nov;32(6):488-496. doi: 10.1097/BOR.0000000000000737. Curr Opin Rheumatol. 2020. PMID: 32941246 Free PMC article. Review.
References
REFERENCES.
METHODS REFERENCES.
-
- Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

