Transcriptome-wide Mapping of Internal N7-Methylguanosine Methylome in Mammalian mRNA
- PMID: 31031084
- PMCID: PMC6588483
- DOI: 10.1016/j.molcel.2019.03.036
Transcriptome-wide Mapping of Internal N7-Methylguanosine Methylome in Mammalian mRNA
Abstract
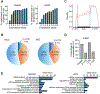
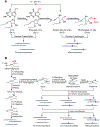
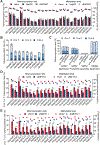
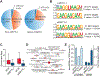
N7-methylguanosine (m7G) is a positively charged, essential modification at the 5' cap of eukaryotic mRNA, regulating mRNA export, translation, and splicing. m7G also occurs internally within tRNA and rRNA, but its existence and distribution within eukaryotic mRNA remain to be investigated. Here, we show the presence of internal m7G sites within mammalian mRNA. We then performed transcriptome-wide profiling of internal m7G methylome using m7G-MeRIP sequencing (MeRIP-seq). To map this modification at base resolution, we developed a chemical-assisted sequencing approach that selectively converts internal m7G sites into abasic sites, inducing misincorporation at these sites during reverse transcription. This base-resolution m7G-seq enabled transcriptome-wide mapping of m7G in human tRNA and mRNA, revealing distribution features of the internal m7G methylome in human cells. We also identified METTL1 as a methyltransferase that installs a subset of m7G within mRNA and showed that internal m7G methylation could affect mRNA translation.
Keywords: METTL1; N(7)-methylguanosine; RNA modification; base-resolution; epitranscriptomics; m(7)G; m(7)G-MeRIP-seq; m(7)G-seq; mRNA modification; tRNA modification; translation regulation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
C.H. is a scientific founder and a scientific advisory board member of Accent Therapeutics. Inc. and a shareholder of Epican Genetech.
Figures


Comment in
-
Put the Pedal to the METTL1: Adding Internal m7G Increases mRNA Translation Efficiency and Augments miRNA Processing.Mol Cell. 2019 Jun 20;74(6):1105-1107. doi: 10.1016/j.molcel.2019.06.004. Mol Cell. 2019. PMID: 31226274
Similar articles
-
Mettl1/Wdr4-Mediated m7G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation.Mol Cell. 2018 Jul 19;71(2):244-255.e5. doi: 10.1016/j.molcel.2018.06.001. Epub 2018 Jul 5. Mol Cell. 2018. PMID: 29983320 Free PMC article.
-
Dynamic methylome of internal mRNA N7-methylguanosine and its regulatory role in translation.Cell Res. 2019 Nov;29(11):927-941. doi: 10.1038/s41422-019-0230-z. Epub 2019 Sep 13. Cell Res. 2019. PMID: 31520064 Free PMC article.
-
Base-Resolution Sequencing Methods for Whole-Transcriptome Quantification of mRNA Modifications.Acc Chem Res. 2024 Jan 2;57(1):47-58. doi: 10.1021/acs.accounts.3c00532. Epub 2023 Dec 11. Acc Chem Res. 2024. PMID: 38079380 Free PMC article.
-
Mapping and significance of the mRNA methylome.Wiley Interdiscip Rev RNA. 2013 Jul-Aug;4(4):397-422. doi: 10.1002/wrna.1166. Epub 2013 May 16. Wiley Interdiscip Rev RNA. 2013. PMID: 23681756 Review.
-
7-Methylguanosine Modifications in Transfer RNA (tRNA).Int J Mol Sci. 2018 Dec 17;19(12):4080. doi: 10.3390/ijms19124080. Int J Mol Sci. 2018. PMID: 30562954 Free PMC article. Review.
Cited by
-
Methylation of Ribosomal RNA: A Mitochondrial Perspective.Front Genet. 2020 Jul 17;11:761. doi: 10.3389/fgene.2020.00761. eCollection 2020. Front Genet. 2020. PMID: 32765591 Free PMC article. Review.
-
Comprehensive pan-cancer analysis of N7-methylguanosine regulators: Expression features and potential implications in prognosis and immunotherapy.Front Genet. 2022 Oct 21;13:1016797. doi: 10.3389/fgene.2022.1016797. eCollection 2022. Front Genet. 2022. PMID: 36339001 Free PMC article.
-
RNA binding proteins in MLL-rearranged leukemia.Exp Hematol Oncol. 2022 Oct 28;11(1):80. doi: 10.1186/s40164-022-00343-5. Exp Hematol Oncol. 2022. PMID: 36307883 Free PMC article. Review.
-
RNA-modifying enzymes and their function in a chromatin context.Nat Struct Mol Biol. 2019 Oct;26(10):858-862. doi: 10.1038/s41594-019-0312-0. Epub 2019 Oct 3. Nat Struct Mol Biol. 2019. PMID: 31582848 Free PMC article. Review.
-
CAP-MAP: cap analysis protocol with minimal analyte processing, a rapid and sensitive approach to analysing mRNA cap structures.Open Biol. 2020 Feb;10(2):190306. doi: 10.1098/rsob.190306. Epub 2020 Feb 26. Open Biol. 2020. PMID: 32097574 Free PMC article.
References
-
- Helm M, and Motorin Y (2017). Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet 18, 275–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

