Biosynthesis of DHGA12 and its roles in Arabidopsis seedling establishment
- PMID: 30992454
- PMCID: PMC6467921
- DOI: 10.1038/s41467-019-09467-5
Biosynthesis of DHGA12 and its roles in Arabidopsis seedling establishment
Abstract
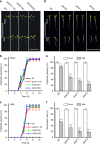
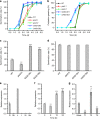
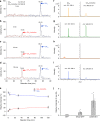
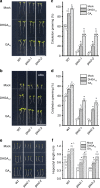
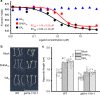
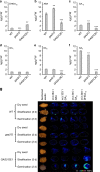
Seed germination and photoautotrophic establishment are controlled by the antagonistic activity of the phytohormones gibberellins (GAs) and abscisic acid (ABA). Here we show that Arabidopsis thaliana GAS2 (Gain of Function in ABA-modulated Seed Germination 2), a protein belonging to the Fe-dependent 2-oxoglutarate dioxygenase superfamily, catalyzes the stereospecific hydration of GA12 to produce GA12 16, 17-dihydro-16α-ol (DHGA12). We show that DHGA12, a C20-GA has an atypical structure compared to known active GAs but can bind to the GA receptor (GID1c). DHGA12 can promote seed germination, hypocotyl elongation and cotyledon greening. Silencing and over-expression of GAS2 alters the ABA/GA ratio and sensitivity to ABA during seed germination and photoautotrophic establishment. Hence, we propose that GAS2 acts to modulate hormonal balance during early seedling development.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
A novel gibberellin promotes seedling establishment.Nat Plants. 2019 May;5(5):459-460. doi: 10.1038/s41477-019-0427-7. Nat Plants. 2019. PMID: 31036915 No abstract available.
Similar articles
-
The dioxygenase GIM2 functions in seed germination by altering gibberellin production in Arabidopsis.J Integr Plant Biol. 2018 Apr;60(4):276-291. doi: 10.1111/jipb.12619. Epub 2018 Feb 24. J Integr Plant Biol. 2018. PMID: 29205921
-
The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels.BMC Plant Biol. 2016 Sep 9;16(1):198. doi: 10.1186/s12870-016-0890-5. BMC Plant Biol. 2016. PMID: 27613195 Free PMC article.
-
AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds.Plant J. 2020 Jan;101(2):310-323. doi: 10.1111/tpj.14542. Epub 2019 Oct 22. Plant J. 2020. PMID: 31536657
-
Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination.Plant Cell Rep. 2012 Feb;31(2):253-70. doi: 10.1007/s00299-011-1180-1. Epub 2011 Nov 2. Plant Cell Rep. 2012. PMID: 22044964 Review.
-
The Multifunctional Dioxygenases of Gibberellin Synthesis.Plant Cell Physiol. 2020 Dec 23;61(11):1869-1879. doi: 10.1093/pcp/pcaa051. Plant Cell Physiol. 2020. PMID: 32343806 Review.
Cited by
-
The Current Status of Research on Gibberellin Biosynthesis.Plant Cell Physiol. 2020 Dec 23;61(11):1832-1849. doi: 10.1093/pcp/pcaa092. Plant Cell Physiol. 2020. PMID: 32652020 Free PMC article. Review.
-
Screening of potential chemical marker with interspecific differences in Pterocarpus wood and a spatially-resolved approach to visualize the distribution of the characteristic markers.Front Plant Sci. 2023 Feb 14;14:1133848. doi: 10.3389/fpls.2023.1133848. eCollection 2023. Front Plant Sci. 2023. PMID: 36866375 Free PMC article.
-
14-3-3 gene of Zostera japonica ZjGRF1 participates in gibberellin signaling pathway.Mol Biol Rep. 2022 Jun;49(6):4795-4803. doi: 10.1007/s11033-022-07330-3. Epub 2022 Apr 15. Mol Biol Rep. 2022. PMID: 35426551
-
Gibberellin Metabolism in Flowering Plants: An Update and Perspectives.Front Plant Sci. 2020 May 19;11:532. doi: 10.3389/fpls.2020.00532. eCollection 2020. Front Plant Sci. 2020. PMID: 32508855 Free PMC article.
-
Manipulating GA-Related Genes for Cereal Crop Improvement.Int J Mol Sci. 2022 Nov 14;23(22):14046. doi: 10.3390/ijms232214046. Int J Mol Sci. 2022. PMID: 36430524 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

