A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis
- PMID: 30962421
- PMCID: PMC6453886
- DOI: 10.1038/s41467-019-09277-9
A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis
Abstract
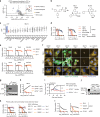
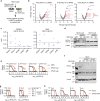
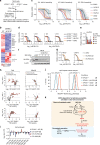
Clear-cell carcinomas (CCCs) are a histological group of highly aggressive malignancies commonly originating in the kidney and ovary. CCCs are distinguished by aberrant lipid and glycogen accumulation and are refractory to a broad range of anti-cancer therapies. Here we identify an intrinsic vulnerability to ferroptosis associated with the unique metabolic state in CCCs. This vulnerability transcends lineage and genetic landscape, and can be exploited by inhibiting glutathione peroxidase 4 (GPX4) with small-molecules. Using CRISPR screening and lipidomic profiling, we identify the hypoxia-inducible factor (HIF) pathway as a driver of this vulnerability. In renal CCCs, HIF-2α selectively enriches polyunsaturated lipids, the rate-limiting substrates for lipid peroxidation, by activating the expression of hypoxia-inducible, lipid droplet-associated protein (HILPDA). Our study suggests targeting GPX4 as a therapeutic opportunity in CCCs, and highlights that therapeutic approaches can be identified on the basis of cell states manifested by morphological and metabolic features in hard-to-treat cancers.
Conflict of interest statement
S.L.S. is a member of the Board of Directors of the Genomics Institute of the Novartis Research Foundation (GNF); a shareholder and member of the Board of Directors of Jnana Therapeutics; a shareholder of Forma Therapeutics; a shareholder of and adviser to Decibel Therapeutics and Eikonizo Therapeutics; an adviser to Eisai, Inc., the Ono Pharma Foundation, and F-Prime Capital Partners; and a Novartis Faculty Scholar. P.A.C. is an adviser to Pfizer, Inc. S. Signoretti has consulting or advisory role for AstraZeneca/MedImmune, Merck, AACR, NCI; royalties from Biogenex Laboratories; and research funding from AstraZeneca, Exelixis, Bristol-Myers Squibb. T.K.C. receives institutional and personal research funds from AstraZeneca, Bayer, BMS, Cerulean, Eisai, Foundation Medicine Inc., Exelixis, Ipsen, Tracon, Genentech, Roche, Roche Products Limited, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, Prometheus Labs, Corvus, Calithera, Analysis Group, Takeda; and receives personal honoraria from AstraZeneca, Alexion, Sanofi/Aventis, Bayer, BMS, Cerulean, Eisai, Foundation Medicine Inc., Exelixis, Genentech, Roche, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, EMD Serono, Prometheus Labs, Corvus, Ipsen, Up-to-Date, NCCN, Analysis Group, NCCN, Michael J. Hennessy (MJH) Associates, Inc (Healthcare Communications Company with several brands such as OnClive and PER), L-path, Kidney Cancer Journal, Clinical Care Options, Platform Q, Navinata Healthcare, Harborside Press, American Society of Medical Oncology, NEJM, Lancet Oncology, Heron Therapeutics; and has consulting or advisory role for AstraZeneca, Alexion, Sanofi/Aventis, Bayer, BMS, Cerulean, Eisai, Foundation Medicine Inc., Exelixis, Genentech, Heron Therapeutics, Roche, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, EMD Serono, Prometheus Labs, Corvus, Ipsen, Up-to-Date, NCCN, Analysis Group. No speaker’s bureau. No leadership or employment in for-profit companies. Other present or past leadership roles for T.K.C.: Director of GU Oncology Division at Dana-Farber and past President of medical Staff at Dana-Farber), member of NCCN Kidney panel and the GU Steering Committee, past chairman of the Kidney cancer Association Medical and Scientific Steering Committee). No Patents, royalties or other intellectual properties. Travel, accommodations, expenses, in relation to consulting, advisory roles, or honoraria. Medical writing and editorial assistance support may have been funded by Communications companies funded by pharmaceutical companies. The institution (Dana-Farber Cancer Institute) may have received additional independent funding of drug companies or/and royalties potentially involved in research around the subject matter. CV provided upon request for scope of clinical practice and research. The remaining authors declare no competing interests.
Figures

Similar articles
-
Identification of CDCP1 as a hypoxia-inducible factor 2α (HIF-2α) target gene that is associated with survival in clear cell renal cell carcinoma patients.Proc Natl Acad Sci U S A. 2013 Feb 26;110(9):3483-8. doi: 10.1073/pnas.1222435110. Epub 2013 Feb 1. Proc Natl Acad Sci U S A. 2013. PMID: 23378636 Free PMC article.
-
The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma.Oncogene. 2018 Oct;37(40):5435-5450. doi: 10.1038/s41388-018-0315-z. Epub 2018 Jun 5. Oncogene. 2018. PMID: 29872221 Free PMC article.
-
HIF-2α expression and metabolic signaling require ACSS2 in clear cell renal cell carcinoma.J Clin Invest. 2024 Jun 17;134(12):e164249. doi: 10.1172/JCI164249. J Clin Invest. 2024. PMID: 38941296 Free PMC article.
-
Hypoxia, Hypoxia-inducible Transcription Factors, and Renal Cancer.Eur Urol. 2016 Apr;69(4):646-657. doi: 10.1016/j.eururo.2015.08.007. Epub 2015 Aug 19. Eur Urol. 2016. PMID: 26298207 Free PMC article. Review.
-
Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis.Curr Top Microbiol Immunol. 2017;403:143-170. doi: 10.1007/82_2016_508. Curr Top Microbiol Immunol. 2017. PMID: 28204974 Review.
Cited by
-
Ferroptosis: Emerging mechanisms, biological function, and therapeutic potential in cancer and inflammation.Cell Death Discov. 2024 Jan 24;10(1):45. doi: 10.1038/s41420-024-01825-7. Cell Death Discov. 2024. PMID: 38267442 Free PMC article. Review.
-
Ferroptosis-Related Gene Signature Accurately Predicts Survival Outcomes in Patients With Clear-Cell Renal Cell Carcinoma.Front Oncol. 2021 Apr 30;11:649347. doi: 10.3389/fonc.2021.649347. eCollection 2021. Front Oncol. 2021. PMID: 33996565 Free PMC article.
-
Targeting AKT induced Ferroptosis through FTO/YTHDF2-dependent GPX4 m6A methylation up-regulating and degradating in colorectal cancer.Cell Death Discov. 2023 Dec 15;9(1):457. doi: 10.1038/s41420-023-01746-x. Cell Death Discov. 2023. PMID: 38102129 Free PMC article.
-
The AMPK-related kinase NUAK2 suppresses glutathione peroxidase 4 expression and promotes ferroptotic cell death in breast cancer cells.Cell Death Discov. 2022 May 6;8(1):253. doi: 10.1038/s41420-022-01044-y. Cell Death Discov. 2022. PMID: 35523770 Free PMC article.
-
Crosstalk between microwave ablation and ferroptosis: The next hot topic?Front Oncol. 2023 Jan 13;13:1099731. doi: 10.3389/fonc.2023.1099731. eCollection 2023. Front Oncol. 2023. PMID: 36712497 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

