Single-Cell Transcriptome Analysis of CD34+ Stem Cell-Derived Myeloid Cells Infected With Human Cytomegalovirus
- PMID: 30949159
- PMCID: PMC6437045
- DOI: 10.3389/fmicb.2019.00577
Single-Cell Transcriptome Analysis of CD34+ Stem Cell-Derived Myeloid Cells Infected With Human Cytomegalovirus
Abstract
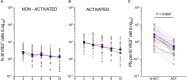
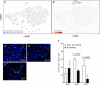
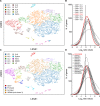
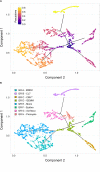
Myeloid cells are important sites of lytic and latent infection by human cytomegalovirus (CMV). We previously showed that only a small subset of myeloid cells differentiated from CD34+ hematopoietic stem cells is permissive to CMV replication, underscoring the heterogeneous nature of these populations. The exact identity of resistant and permissive cell types, and the cellular features characterizing the latter, however, could not be dissected using averaging transcriptional analysis tools such as microarrays and, hence, remained enigmatic. Here, we profile the transcriptomes of ∼7000 individual cells at day 1 post-infection using the 10× genomics platform. We show that viral transcripts are detectable in the majority of the cells, suggesting that virion entry is unlikely to be the main target of cellular restriction mechanisms. We further show that viral replication occurs in a small but specific sub-group of cells transcriptionally related to, and likely derived from, a cluster of cells expressing markers of Colony Forming Unit - Granulocyte, Erythrocyte, Monocyte, Megakaryocyte (CFU-GEMM) oligopotent progenitors. Compared to the remainder of the population, CFU-GEMM cells are enriched in transcripts with functions in mitochondrial energy production, cell proliferation, RNA processing and protein synthesis, and express similar or higher levels of interferon-related genes. While expression levels of the former are maintained in infected cells, the latter are strongly down-regulated. We thus propose that the preferential infection of CFU-GEMM cells may be due to the presence of a pre-established pro-viral environment, requiring minimal optimization efforts from viral effectors, rather than to the absence of specific restriction factors. Together, these findings identify a potentially new population of myeloid cells permissive to CMV replication, and provide a possible rationale for their preferential infection.
Keywords: 10× genomics; RNAseq; cytomegalovirus; myeloid; single-cell; tropism.
Figures
Similar articles
-
Dynamics of Human Cytomegalovirus Infection in CD34+ Hematopoietic Cells and Derived Langerhans-Type Dendritic Cells.J Virol. 2015 May;89(10):5615-32. doi: 10.1128/JVI.00305-15. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762731 Free PMC article.
-
Cytokine-dependent ex vivo expansion of early subsets of CD34+ cord blood myeloid progenitors is enhanced by cord blood plasma, but expansion of the more mature subsets of progenitors is favored.Blood Cells. 1994;20(2-3):436-54. Blood Cells. 1994. PMID: 7538350
-
Interleukin-11 stimulates the proliferation of human hematopoietic CD34+ and CD34+CD33-DR- cells and synergizes with stem cell factor, interleukin-3, and granulocyte-macrophage colony-stimulating factor.Exp Hematol. 1993 Dec;21(13):1668-72. Exp Hematol. 1993. PMID: 7694867
-
Human cytomegalovirus latent infection of granulocyte-macrophage progenitors.Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11879-83. doi: 10.1073/pnas.91.25.11879. Proc Natl Acad Sci U S A. 1994. PMID: 7991550 Free PMC article.
-
HCMV Displays a Unique Transcriptome of Immunomodulatory Genes in Primary Monocyte-Derived Cell Types.PLoS One. 2016 Oct 19;11(10):e0164843. doi: 10.1371/journal.pone.0164843. eCollection 2016. PLoS One. 2016. PMID: 27760232 Free PMC article.
Cited by
-
Cytomegalovirus Latency and Reactivation: An Intricate Interplay With the Host Immune Response.Front Cell Infect Microbiol. 2020 Mar 31;10:130. doi: 10.3389/fcimb.2020.00130. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32296651 Free PMC article. Review.
-
Regulation of the MIE Locus During HCMV Latency and Reactivation.Pathogens. 2020 Oct 23;9(11):869. doi: 10.3390/pathogens9110869. Pathogens. 2020. PMID: 33113934 Free PMC article. Review.
-
Single-Cell RNA-seq Identifies Cell Subsets in Human Placenta That Highly Expresses Factors Driving Pathogenesis of SARS-CoV-2.Front Cell Dev Biol. 2020 Aug 19;8:783. doi: 10.3389/fcell.2020.00783. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32974340 Free PMC article.
-
Human cytomegalovirus and neonatal infection.Curr Res Microb Sci. 2024 Jun 24;7:100257. doi: 10.1016/j.crmicr.2024.100257. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39070527 Free PMC article. Review.
-
Understanding HCMV Latency Using Unbiased Proteomic Analyses.Pathogens. 2020 Jul 20;9(7):590. doi: 10.3390/pathogens9070590. Pathogens. 2020. PMID: 32698381 Free PMC article. Review.
References
-
- Biolatti M., Dell’Oste V., Pautasso S., Gugliesi F., von Einem J., Krapp C., et al. (2018). Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J. Virol. 92:e01774-17 10.1128/JVI.01774-17 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

