Histone Deacetylase Inhibitor Suberoylanilide Hydroxamic Acid Suppresses Human Adenovirus Gene Expression and Replication
- PMID: 30944181
- PMCID: PMC6613751
- DOI: 10.1128/JVI.00088-19
Histone Deacetylase Inhibitor Suberoylanilide Hydroxamic Acid Suppresses Human Adenovirus Gene Expression and Replication
Abstract
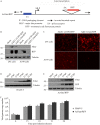
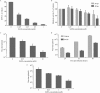
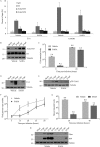
Human adenovirus (HAdV) causes minor illnesses in most patients but can lead to severe disease and death in pediatric, geriatric, and immunocompromised individuals. No approved antiviral therapy currently exists for the treatment of these severe HAdV-induced diseases. In this study, we show that the pan-histone deacetylase (HDAC) inhibitor SAHA reduces HAdV-5 gene expression and DNA replication in tissue culture, ultimately decreasing virus yield from infected cells. Importantly, SAHA also reduced gene expression from more virulent and clinically relevant serotypes, including HAdV-4 and HAdV-7. In addition to SAHA, several other HDAC inhibitors (e.g., trichostatin A, apicidin, and panobinostat) also affected HAdV gene expression. We determined that loss of class I HDAC activity, mainly HDAC2, impairs efficient expression of viral genes, and that E1A physically interacts with HDAC2. Our results suggest that HDAC activity is necessary for HAdV replication, which may represent a novel pharmacological target in HAdV-induced disease.IMPORTANCE Although human adenovirus (HAdV) can cause severe diseases that can be fatal in some populations, there are no effective treatments to combat HAdV infection. In this study, we determined that the pan-histone deacetylase (HDAC) inhibitor SAHA has inhibitory activity against several clinically relevant serotypes of HAdV. This U.S. Food and Drug Administration-approved compound affects various stages of the virus lifecycle and reduces virus yield even at low concentrations. We further report that class I HDAC activity, particularly HDAC2, is required for efficient expression of viral genes during lytic infection. Investigation of the mechanism underlying SAHA-mediated suppression of HAdV gene expression and replication will enhance current knowledge of virus-cell interaction and may aid in the development of more effective antivirals with lower toxicity for the treatment of HAdV infections.
Keywords: SAHA; adenoviruses; histone deacetylase; histone deacetylase inhibitors; vorinostat.
Copyright © 2019 American Society for Microbiology.
Figures





Similar articles
-
Histone Deacetylase Inhibitors Promote Latent Adenovirus Reactivation from Tonsillectomy Specimens.J Virol. 2020 Jun 1;94(12):e00100-20. doi: 10.1128/JVI.00100-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32269118 Free PMC article.
-
The histone deacetylase inhibitor vorinostat (SAHA) increases the susceptibility of uninfected CD4+ T cells to HIV by increasing the kinetics and efficiency of postentry viral events.J Virol. 2014 Sep;88(18):10803-12. doi: 10.1128/JVI.00320-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008921 Free PMC article.
-
Histone deacetylase inhibition is cytotoxic to oligodendrocyte precursor cells in vitro and in vivo.Int J Dev Neurosci. 2016 Nov;54:53-61. doi: 10.1016/j.ijdevneu.2016.08.006. Epub 2016 Aug 29. Int J Dev Neurosci. 2016. PMID: 27587342
-
Suberoylanilide Hydroxamic Acid Analogs with Heteroaryl Amide Group and Different Chain Length: Synthesis and Effect on Histone Deacetylase.Molecules. 2024 Jan 1;29(1):238. doi: 10.3390/molecules29010238. Molecules. 2024. PMID: 38202821 Free PMC article. Review.
-
Inhibitors of histone deacetylase as antitumor agents: A critical review.Bioorg Chem. 2016 Aug;67:18-42. doi: 10.1016/j.bioorg.2016.05.005. Epub 2016 May 17. Bioorg Chem. 2016. PMID: 27239721 Review.
Cited by
-
HDAC Inhibitors Enhance Efficacy of the Oncolytic Adenoviruses Ad∆∆ and Ad-3∆-A20T in Pancreatic and Triple-Negative Breast Cancer Models.Viruses. 2022 May 9;14(5):1006. doi: 10.3390/v14051006. Viruses. 2022. PMID: 35632748 Free PMC article.
-
Antiviral Effects of Curcumin on Adenovirus Replication.Microorganisms. 2020 Oct 4;8(10):1524. doi: 10.3390/microorganisms8101524. Microorganisms. 2020. PMID: 33020422 Free PMC article.
-
Respiratory illness virus infections with special emphasis on COVID-19.Eur J Med Res. 2022 Nov 8;27(1):236. doi: 10.1186/s40001-022-00874-x. Eur J Med Res. 2022. PMID: 36348452 Free PMC article. Review.
-
HDAC Inhibition as Potential Therapeutic Strategy to Restore the Deregulated Immune Response in Severe COVID-19.Front Immunol. 2022 May 3;13:841716. doi: 10.3389/fimmu.2022.841716. eCollection 2022. Front Immunol. 2022. PMID: 35592335 Free PMC article.
-
Expanding the adenovirus toolbox: reporter viruses for studying the dynamics of human adenovirus replication.J Virol. 2024 May 14;98(5):e0020724. doi: 10.1128/jvi.00207-24. Epub 2024 Apr 19. J Virol. 2024. PMID: 38639487 Free PMC article.
References
-
- Ying B, Tollefson AE, Spencer JF, Balakrishnan L, Dewhurst S, Capella C, Buller RM, Toth K, Wold WS. 2014. Ganciclovir inhibits human adenovirus replication and pathogenicity in permissive immunosuppressed Syrian hamsters. Antimicrob Agents Chemother 58:7171–7181. doi:10.1128/AAC.03860-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

