Point mutations in the PDX1 transactivation domain impair human β-cell development and function
- PMID: 30930126
- PMCID: PMC6531841
- DOI: 10.1016/j.molmet.2019.03.006
Point mutations in the PDX1 transactivation domain impair human β-cell development and function
Abstract
Objective: Hundreds of missense mutations in the coding region of PDX1 exist; however, if these mutations predispose to diabetes mellitus is unknown.
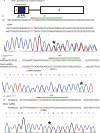
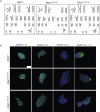
Methods: In this study, we screened a large cohort of subjects with increased risk for diabetes and identified two subjects with impaired glucose tolerance carrying common, heterozygous, missense mutations in the PDX1 coding region leading to single amino acid exchanges (P33T, C18R) in its transactivation domain. We generated iPSCs from patients with heterozygous PDX1P33T/+, PDX1C18R/+ mutations and engineered isogenic cell lines carrying homozygous PDX1P33T/P33T, PDX1C18R/C18R mutations and a heterozygous PDX1 loss-of-function mutation (PDX1+/-).
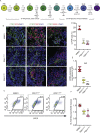
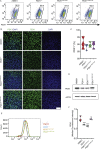
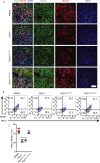
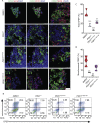
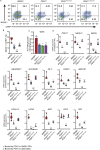
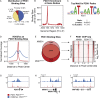
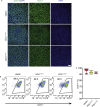
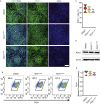
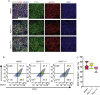
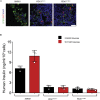
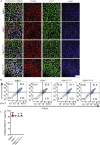
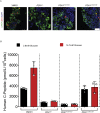
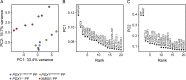
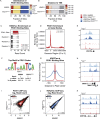
Results: Using an in vitro β-cell differentiation protocol, we demonstrated that both, heterozygous PDX1P33T/+, PDX1C18R/+ and homozygous PDX1P33T/P33T, PDX1C18R/C18R mutations impair β-cell differentiation and function. Furthermore, PDX1+/- and PDX1P33T/P33T mutations reduced differentiation efficiency of pancreatic progenitors (PPs), due to downregulation of PDX1-bound genes, including transcription factors MNX1 and PDX1 as well as insulin resistance gene CES1. Additionally, both PDX1P33T/+ and PDX1P33T/P33T mutations in PPs reduced the expression of PDX1-bound genes including the long-noncoding RNA, MEG3 and the imprinted gene NNAT, both involved in insulin synthesis and secretion.
Conclusions: Our results reveal mechanistic details of how common coding mutations in PDX1 impair human pancreatic endocrine lineage formation and β-cell function and contribute to the predisposition for diabetes.
Keywords: Insulin secretion; PDX1; PDX1-Bound genes; Transactivation domain; β-Cell differentiation.
Copyright © 2019 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures



Similar articles
-
Genome-wide analysis of PDX1 target genes in human pancreatic progenitors.Mol Metab. 2018 Mar;9:57-68. doi: 10.1016/j.molmet.2018.01.011. Epub 2018 Jan 31. Mol Metab. 2018. PMID: 29396371 Free PMC article.
-
Generation of a human induced pluripotent stem cell (iPSC) line from a patient carrying a P33T mutation in the PDX1 gene.Stem Cell Res. 2016 Sep;17(2):273-276. doi: 10.1016/j.scr.2016.08.004. Epub 2016 Aug 5. Stem Cell Res. 2016. PMID: 27879211
-
Generation of a human induced pluripotent stem cell (iPSC) line from a patient with family history of diabetes carrying a C18R mutation in the PDX1 gene.Stem Cell Res. 2016 Sep;17(2):292-295. doi: 10.1016/j.scr.2016.08.005. Epub 2016 Aug 6. Stem Cell Res. 2016. PMID: 27879214
-
PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration.Stem Cell Res Ther. 2017 Nov 2;8(1):240. doi: 10.1186/s13287-017-0694-z. Stem Cell Res Ther. 2017. PMID: 29096722 Free PMC article. Review.
-
Unlocking β-cell restoration: The crucial role of PDX1 in diabetes therapy.Pathol Res Pract. 2024 Feb;254:155131. doi: 10.1016/j.prp.2024.155131. Epub 2024 Jan 20. Pathol Res Pract. 2024. PMID: 38309018 Review.
Cited by
-
Transcriptome-Powered Pluripotent Stem Cell Differentiation for Regenerative Medicine.Cells. 2023 May 22;12(10):1442. doi: 10.3390/cells12101442. Cells. 2023. PMID: 37408278 Free PMC article. Review.
-
What Do We Know about Neonatal Diabetes caused by PDX1 Mutations?Curr Diabetes Rev. 2024;21(1):e290124226471. doi: 10.2174/0115733998265866231204070606. Curr Diabetes Rev. 2024. PMID: 38299270 Review.
-
The Role of ER Stress in Diabetes: Exploring Pathological Mechanisms Using Wolfram Syndrome.Int J Mol Sci. 2022 Dec 23;24(1):230. doi: 10.3390/ijms24010230. Int J Mol Sci. 2022. PMID: 36613674 Free PMC article. Review.
-
Dynamic regulation of pancreatic β cell function and gene expression by the SND1 coregulator in vitro.Islets. 2023 Dec 31;15(1):2267725. doi: 10.1080/19382014.2023.2267725. Epub 2023 Oct 15. Islets. 2023. PMID: 37838950 Free PMC article.
-
Gene-edited human stem cell-derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice.Sci Transl Med. 2020 Apr 22;12(540):eaax9106. doi: 10.1126/scitranslmed.aax9106. Sci Transl Med. 2020. PMID: 32321868 Free PMC article.
References
-
- Ahlqvist E., Storm P., Käräjämäki A., Martinell M., Dorkhan M., Carlsson A. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. The Lancet Diabetes and Endocrinology. 2018;6(5):361–369. - PubMed
-
- Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014;37(Supplement_1):S81–S90. - PubMed
-
- Staffers D.A., Ferrer J., Clarke W.L., Habener J.F. Early-onset type-ll diabetes mellitus (MODY4) linked to IPF1. Nature Genetics. 1997;17(2):138–139. - PubMed
-
- Heuvel-Borsboom H., de Valk H.W., Losekoot M., Westerink J. Maturity onset diabetes of the young: seek and you will find. The Netherlands Journal of Medicine. 2016:193–200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

