Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles
- PMID: 30872707
- PMCID: PMC6418312
- DOI: 10.1038/s41598-019-41029-z
Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles
Abstract
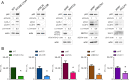
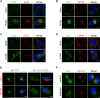
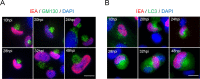
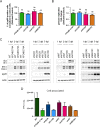
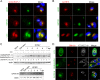
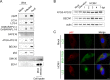
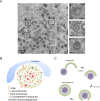
During its life cycle, Human cytomegalovirus (HCMV) tightly modulates autophagy, a vesicular pathway allowing degradation and recycling of cellular components. To study the interplay between autophagy and the viral life cycle, we established various autophagy-deficient human fibroblastic cell lines. By knocking down the expression or activity of five autophagy-related proteins, we confirmed the proviral function that the autophagic machinery exerts on HCMV production. Using 3D reconstruction from confocal microscopy and electron microscopy, we demonstrated that lipidated LC3-positive vesicles accumulated at the viral assembly compartment (vAC). The vAC is a juxtanuclear ring-shaped structure containing several organelles and membranes, where assembly and final envelopment of HCMV particles occur. Two LC3 homologs, GABARAPL1 and GATE16, also accumulated during HCMV infection and were associated with the vAC, in proximity with fragmented Golgi stacks. Additionally, we observed the formation of a pre-assembly compartment (PrAC) in infected cells, which consists of a juxtanuclear structure containing both fragmented Golgi and LC3-positive vesicles. Finally, we showed that highly purified extracellular viral particles were associated with various autophagy proteins. Our results thus suggest that autophagy machinery participates to the final cytoplasmic envelopment of HCMV viral particles into the vAC and that autophagy-related proteins can be spotted in the virions.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Human Cytomegalovirus Hijacks WD Repeat Domain 11 for Virion Assembly Compartment Formation and Virion Morphogenesis.J Virol. 2022 Mar 9;96(5):e0182721. doi: 10.1128/JVI.01827-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35020472 Free PMC article.
-
Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex.J Virol. 2014 Aug;88(16):9086-99. doi: 10.1128/JVI.01141-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899189 Free PMC article.
-
Phosphorylation of Golgi Peripheral Membrane Protein Grasp65 Is an Integral Step in the Formation of the Human Cytomegalovirus Cytoplasmic Assembly Compartment.mBio. 2016 Oct 4;7(5):e01554-16. doi: 10.1128/mBio.01554-16. mBio. 2016. PMID: 27703074 Free PMC article.
-
Host Cell Signatures of the Envelopment Site within Beta-Herpes Virions.Int J Mol Sci. 2022 Sep 1;23(17):9994. doi: 10.3390/ijms23179994. Int J Mol Sci. 2022. PMID: 36077391 Free PMC article. Review.
-
Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy.BMB Rep. 2016 Aug;49(8):424-30. doi: 10.5483/bmbrep.2016.49.8.081. BMB Rep. 2016. PMID: 27418283 Free PMC article. Review.
Cited by
-
Modulation of Endosome Function, Vesicle Trafficking and Autophagy by Human Herpesviruses.Cells. 2021 Mar 4;10(3):542. doi: 10.3390/cells10030542. Cells. 2021. PMID: 33806291 Free PMC article. Review.
-
Human Cytomegalovirus Modifies Placental Small Extracellular Vesicle Composition to Enhance Infection of Fetal Neural Cells In Vitro.Viruses. 2022 Sep 13;14(9):2030. doi: 10.3390/v14092030. Viruses. 2022. PMID: 36146834 Free PMC article.
-
Secretory autophagy promotes RAB37-mediated insulin secretion under glucose stimulation both in vitro and in vivo.Autophagy. 2023 Apr;19(4):1239-1257. doi: 10.1080/15548627.2022.2123098. Epub 2022 Sep 15. Autophagy. 2023. PMID: 36109708 Free PMC article.
-
Cell type-specific biogenesis of novel vesicles containing viral products in human cytomegalovirus infection.J Virol. 2021 May 10;95(11):e02358-20. doi: 10.1128/JVI.02358-20. Epub 2021 Mar 24. J Virol. 2021. PMID: 33762413 Free PMC article.
-
Aspirin and 5-Aminoimidazole-4-carboxamide Riboside Attenuate Bovine Ephemeral Fever Virus Replication by Inhibiting BEFV-Induced Autophagy.Front Immunol. 2020 Nov 24;11:556838. doi: 10.3389/fimmu.2020.556838. eCollection 2020. Front Immunol. 2020. PMID: 33329515 Free PMC article.
References
-
- Mocarski, E. S., Shenk, T., Griffiths, P. & Pass, R. F. In Fields Virology, 6th ed. Vol. vol2 (eds Knipe, D. M. & Howley, P. M.) (Wolters Kluwer, Lippincott Williams & Wilkins, 2013).

