Human Cytomegalovirus DNA Polymerase Subunit UL44 Antagonizes Antiviral Immune Responses by Suppressing IRF3- and NF-κB-Mediated Transcription
- PMID: 30867312
- PMCID: PMC6532097
- DOI: 10.1128/JVI.00181-19
Human Cytomegalovirus DNA Polymerase Subunit UL44 Antagonizes Antiviral Immune Responses by Suppressing IRF3- and NF-κB-Mediated Transcription
Abstract
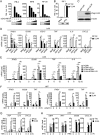
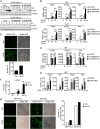
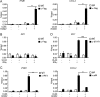
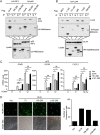
Innate immunity is the first line of host defense against viral invasion. The induction of type I interferons (IFNs) and inflammatory cytokines is essential to host antiviral immune responses, which are also key targets of viral immune evasion. Human cytomegalovirus (HCMV) can establish long-term latent infections, in which immune evasion is a pivotal step. In this study, we identified HCMV protein UL44, a DNA polymerase processivity factor, as an inhibitor of the interferon regulatory factor 3 (IRF3)- and NF-κB-dependent antiviral response. Ectopic expression of UL44 inhibited HCMV-triggered induction of downstream effector genes and enhanced viral replication. Conversely, knockdown of UL44 potentiated HCMV-triggered induction of downstream antiviral genes. UL44 interacted with IRF3 and p65, and it inhibited the binding of IRF3 and NF-κB to the promoters of their downstream antiviral genes. These findings reveal an important mechanism of immune evasion by HCMV at the transcriptional level.IMPORTANCE Induction of type I IFNs and inflammatory cytokines plays pivotal roles in host antiviral innate immune responses. Viruses have evolved various mechanisms to interfere with these processes. HCMV causes severe ailments in immunodeficient populations and is a major cause of birth defects. It has been shown that HCMV antagonizes host innate immune defenses, which is important for establishing immune evasion and latent infection. In this study, we identified the HCMV DNA polymerase subunit UL44 as a suppressor of antiviral innate immune responses. Overexpression of UL44 impaired HCMV-triggered induction of type I IFNs and other antiviral genes and thus potentiated viral replication, whereas UL44 deficiency showed opposite effects. Mechanistic studies indicated that UL44 acts by inhibiting the binding of IRF3 and NF-κB to the promoters of downstream antiviral genes. These findings defined an important mechanism of HCMV immune evasion at the transcriptional level, which may provide a therapeutic target for the treatment of HCMV infection.
Keywords: HCMV; IRF3; NF-κB; UL44; immune evasion; innate immunity; type I interferons.
Copyright © 2019 American Society for Microbiology.
Figures

Similar articles
-
Human Cytomegalovirus Protein UL94 Targets MITA to Evade the Antiviral Immune Response.J Virol. 2020 Jun 1;94(12):e00022-20. doi: 10.1128/JVI.00022-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32238587 Free PMC article.
-
Induction of INKIT by Viral Infection Negatively Regulates Antiviral Responses through Inhibiting Phosphorylation of p65 and IRF3.Cell Host Microbe. 2017 Jul 12;22(1):86-98.e4. doi: 10.1016/j.chom.2017.06.013. Cell Host Microbe. 2017. PMID: 28704656
-
Herpes Simplex Virus 1 Serine Protease VP24 Blocks the DNA-Sensing Signal Pathway by Abrogating Activation of Interferon Regulatory Factor 3.J Virol. 2016 May 27;90(12):5824-5829. doi: 10.1128/JVI.00186-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27076640 Free PMC article.
-
Identification and utility of innate immune system evasion mechanisms of ASFV.Virus Res. 2013 Apr;173(1):87-100. doi: 10.1016/j.virusres.2012.10.013. Epub 2012 Nov 16. Virus Res. 2013. PMID: 23165138 Review.
-
The tiers and dimensions of evasion of the type I interferon response by human cytomegalovirus.J Mol Biol. 2013 Dec 13;425(24):4857-71. doi: 10.1016/j.jmb.2013.08.023. Epub 2013 Sep 5. J Mol Biol. 2013. PMID: 24013068 Free PMC article. Review.
Cited by
-
Of Keeping and Tipping the Balance: Host Regulation and Viral Modulation of IRF3-Dependent IFNB1 Expression.Viruses. 2020 Jul 7;12(7):733. doi: 10.3390/v12070733. Viruses. 2020. PMID: 32645843 Free PMC article. Review.
-
Insights into the Transcriptome of Human Cytomegalovirus: A Comprehensive Review.Viruses. 2023 Aug 8;15(8):1703. doi: 10.3390/v15081703. Viruses. 2023. PMID: 37632045 Free PMC article. Review.
-
The Cytomegalovirus M35 Protein Directly Binds to the Interferon-β Enhancer and Modulates Transcription of Ifnb1 and Other IRF3-Driven Genes.J Virol. 2023 Jun 29;97(6):e0040023. doi: 10.1128/jvi.00400-23. Epub 2023 Jun 8. J Virol. 2023. PMID: 37289084 Free PMC article.
-
SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response.Cell Mol Immunol. 2021 Mar;18(3):613-620. doi: 10.1038/s41423-020-00571-x. Epub 2020 Oct 27. Cell Mol Immunol. 2021. PMID: 33110251 Free PMC article.
-
Placental Immune Responses to Viruses: Molecular and Histo-Pathologic Perspectives.Int J Mol Sci. 2021 Mar 13;22(6):2921. doi: 10.3390/ijms22062921. Int J Mol Sci. 2021. PMID: 33805739 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

