Differences in the path to exit the ribosome across the three domains of life
- PMID: 30805621
- PMCID: PMC6486554
- DOI: 10.1093/nar/gkz106
Differences in the path to exit the ribosome across the three domains of life
Abstract
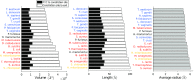
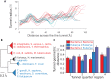
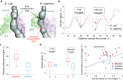
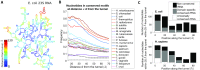
The ribosome exit tunnel is an important structure involved in the regulation of translation and other essential functions such as protein folding. By comparing 20 recently obtained cryo-EM and X-ray crystallography structures of the ribosome from all three domains of life, we here characterize the key similarities and differences of the tunnel across species. We first show that a hierarchical clustering of tunnel shapes closely reflects the species phylogeny. Then, by analyzing the ribosomal RNAs and proteins, we explain the observed geometric variations and show direct association between the conservations of the geometry, structure and sequence. We find that the tunnel is more conserved in the upper part close to the polypeptide transferase center, while in the lower part, it is substantially narrower in eukaryotes than in bacteria. Furthermore, we provide evidence for the existence of a second constriction site in eukaryotic exit tunnels. Overall, these results have several evolutionary and functional implications, which explain certain differences between eukaryotes and prokaryotes in their translation mechanisms. In particular, they suggest that major co-translational functions of bacterial tunnels were externalized in eukaryotes, while reducing the tunnel size provided some other advantages, such as facilitating the nascent chain elongation and enabling antibiotic resistance.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
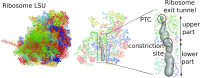


Similar articles
-
Ribosomal proteins: structure, function, and evolution.Biochemistry (Mosc). 2012 Jun;77(6):562-74. doi: 10.1134/S0006297912060028. Biochemistry (Mosc). 2012. PMID: 22817455 Review.
-
Protein Fold Usages in Ribosomes: Another Glance to the Past.Int J Mol Sci. 2024 Aug 13;25(16):8806. doi: 10.3390/ijms25168806. Int J Mol Sci. 2024. PMID: 39201491 Free PMC article.
-
The ribosome in focus.Cell. 2001 Mar 23;104(6):813-6. doi: 10.1016/s0092-8674(01)00278-1. Cell. 2001. PMID: 11290319 Review. No abstract available.
-
Residue conservation elucidates the evolution of r-proteins in ribosomal assembly and function.Int J Biol Macromol. 2019 Nov 1;140:323-329. doi: 10.1016/j.ijbiomac.2019.08.127. Epub 2019 Aug 15. Int J Biol Macromol. 2019. PMID: 31421176
-
Dynamic association of human Ebp1 with the ribosome.RNA. 2021 Apr;27(4):411-419. doi: 10.1261/rna.077602.120. Epub 2021 Jan 21. RNA. 2021. PMID: 33479117 Free PMC article.
Cited by
-
Nascent peptide-induced translation discontinuation in eukaryotes impacts biased amino acid usage in proteomes.Nat Commun. 2022 Dec 2;13(1):7451. doi: 10.1038/s41467-022-35156-x. Nat Commun. 2022. PMID: 36460666 Free PMC article.
-
A Comparative Perspective on Ribosome Biogenesis: Unity and Diversity Across the Tree of Life.Methods Mol Biol. 2022;2533:3-22. doi: 10.1007/978-1-0716-2501-9_1. Methods Mol Biol. 2022. PMID: 35796979 Free PMC article. Review.
-
Comprehensive Transcriptome Analysis Reveals Genome-Wide Changes Associated with Endoplasmic Reticulum (ER) Stress in Potato (Solanum tuberosum L.).Int J Mol Sci. 2022 Nov 9;23(22):13795. doi: 10.3390/ijms232213795. Int J Mol Sci. 2022. PMID: 36430273 Free PMC article.
-
The Identity of the Constriction Region of the Ribosomal Exit Tunnel Is Important to Maintain Gene Expression in Escherichia coli.Microbiol Spectr. 2022 Apr 27;10(2):e0226121. doi: 10.1128/spectrum.02261-21. Epub 2022 Mar 21. Microbiol Spectr. 2022. PMID: 35311583 Free PMC article.
-
Context-specific action of macrolide antibiotics on the eukaryotic ribosome.Nat Commun. 2021 May 14;12(1):2803. doi: 10.1038/s41467-021-23068-1. Nat Commun. 2021. PMID: 33990576 Free PMC article.
References
-
- Nyathi Y., Wilkinson B.M., Pool M.R.. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta. 2013; 1833:2392–2402. - PubMed
-
- Thommen M., Holtkamp W., Rodnina M.V.. Co-translational protein folding: progress and methods. Curr. Opin. Struct. Biol. 2017; 42:83–89. - PubMed
-
- Rodnina M.V., Wintermeyer W.. Protein elongation, co-translational folding and targeting. J. Mol. Biol. 2016; 428:2165–2185. - PubMed
-
- Vazquez-Laslop N., Thum C., Mankin A.S.. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008; 30:190–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

