Autophagic cell death restricts chromosomal instability during replicative crisis
- PMID: 30675059
- PMCID: PMC6557118
- DOI: 10.1038/s41586-019-0885-0
Autophagic cell death restricts chromosomal instability during replicative crisis
Abstract
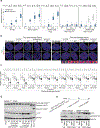
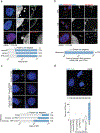
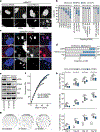
Replicative crisis is a senescence-independent process that acts as a final barrier against oncogenic transformation by eliminating pre-cancerous cells with disrupted cell cycle checkpoints1. It functions as a potent tumour suppressor and culminates in extensive cell death. Cells rarely evade elimination and evolve towards malignancy, but the mechanisms that underlie cell death in crisis are not well understood. Here we show that macroautophagy has a dominant role in the death of fibroblasts and epithelial cells during crisis. Activation of autophagy is critical for cell death, as its suppression promoted bypass of crisis, continued proliferation and accumulation of genome instability. Telomere dysfunction specifically triggers autophagy, implicating a telomere-driven autophagy pathway that is not induced by intrachromosomal breaks. Telomeric DNA damage generates cytosolic DNA species with fragile nuclear envelopes that undergo spontaneous disruption. The cytosolic chromatin fragments activate the cGAS-STING (cyclic GMP-AMP synthase-stimulator of interferon genes) pathway and engage the autophagy machinery. Our data suggest that autophagy is an integral component of the tumour suppressive crisis mechanism and that loss of autophagy function is required for the initiation of cancer.
Figures









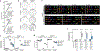

Comment in
-
Telomere crisis activates autophagic death.Nat Rev Mol Cell Biol. 2019 Mar;20(3):133. doi: 10.1038/s41580-019-0105-7. Nat Rev Mol Cell Biol. 2019. PMID: 30700812 No abstract available.
-
Expanding the Role of STING in Cellular Homeostasis and Transformation.Trends Cancer. 2019 Apr;5(4):195-197. doi: 10.1016/j.trecan.2019.02.001. Epub 2019 Feb 23. Trends Cancer. 2019. PMID: 30961826 Free PMC article.
Similar articles
-
Telomeres: Implications for Cancer Development.Int J Mol Sci. 2018 Jan 19;19(1):294. doi: 10.3390/ijms19010294. Int J Mol Sci. 2018. PMID: 29351238 Free PMC article. Review.
-
Cell death during crisis is mediated by mitotic telomere deprotection.Nature. 2015 Jun 25;522(7557):492-6. doi: 10.1038/nature14513. Nature. 2015. PMID: 26108857 Free PMC article.
-
Autolysosomal degradation of cytosolic chromatin fragments antagonizes oxidative stress-induced senescence.J Biol Chem. 2020 Apr 3;295(14):4451-4463. doi: 10.1074/jbc.RA119.010734. Epub 2020 Feb 11. J Biol Chem. 2020. PMID: 32047109 Free PMC article.
-
Molecular mechanisms and cellular functions of cGAS-STING signalling.Nat Rev Mol Cell Biol. 2020 Sep;21(9):501-521. doi: 10.1038/s41580-020-0244-x. Epub 2020 May 18. Nat Rev Mol Cell Biol. 2020. PMID: 32424334 Review.
-
Breast primary epithelial cells that escape p16-dependent stasis enter a telomere-driven crisis state.Breast Cancer Res. 2016 Jan 13;18(1):7. doi: 10.1186/s13058-015-0667-z. Breast Cancer Res. 2016. PMID: 26758019 Free PMC article.
Cited by
-
The influence of circular RNAs on autophagy and disease progression.Autophagy. 2022 Feb;18(2):240-253. doi: 10.1080/15548627.2021.1917131. Epub 2021 Apr 27. Autophagy. 2022. PMID: 33904341 Free PMC article. Review.
-
CagA and VacA inhibit gastric mucosal epithelial cell autophagy and promote the progression of gastric precancerous lesions.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022 Jul 28;47(7):942-951. doi: 10.11817/j.issn.1672-7347.2022.210779. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022. PMID: 36039592 Free PMC article. Chinese, English.
-
Construction of a Prognostic Nomogram Based on Autophagy-Related Genes for Children With Neuroblastoma.Evol Bioinform Online. 2022 Aug 26;18:11769343221120960. doi: 10.1177/11769343221120960. eCollection 2022. Evol Bioinform Online. 2022. PMID: 36046056 Free PMC article.
-
Autophagy-related prognostic signature characterizes tumor microenvironment and predicts response to ferroptosis in gastric cancer.Front Oncol. 2022 Aug 16;12:959337. doi: 10.3389/fonc.2022.959337. eCollection 2022. Front Oncol. 2022. PMID: 36052243 Free PMC article.
-
Role of micronucleus-activated cGAS-STING signaling in antitumor immunity.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024 Jan 26;53(1):25-34. doi: 10.3724/zdxbyxb-2023-0485. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38273467 Free PMC article. Review. Chinese, English.
References
-
- Wei W & Sedivy JM Differentiation between senescence (M1) and crisis (M2) in human fibroblast cultures. Exp. Cell Res. 253, 519–522 (1999). - PubMed
-
- Artandi SE & DePinho RA A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10, 39–46 (2000). - PubMed
-
- Steinberg ML & Defendi V Transformation and immortalization of human keratinocytes by SV40. J. Invest. Dermatol. 81, 131s–136s (1983). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

