Cryo-EM Structure (4.5-Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance
- PMID: 30659798
- PMCID: PMC6378684
- DOI: 10.1016/j.jmb.2019.01.011
Cryo-EM Structure (4.5-Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance
Erratum in
-
Corrigendum to "Cryo-EM Structure (4.5 Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance" [J. Mol. Biol. 431 (2019) 864-872] https://doi.org/10.1016/j.jmb.2019.01.011.J Mol Biol. 2020 Jan 17;432(2):632. doi: 10.1016/j.jmb.2019.08.015. Epub 2019 Nov 15. J Mol Biol. 2020. PMID: 31740076 Free PMC article. No abstract available.
Abstract
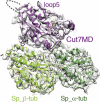
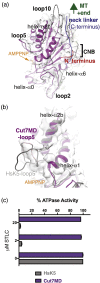
Kinesin-5s are microtubule-dependent motors that drive spindle pole separation during mitosis. We used cryo-electron microscopy to determine the 4.5-Å resolution structure of the motor domain of the fission yeast kinesin-5 Cut7 bound to fission yeast microtubules and explored the topology of the motor-microtubule interface and the susceptibility of the complex to drug binding. Despite their non-canonical architecture and mechanochemistry, Schizosaccharomyces pombe microtubules were stabilized by epothilone at the taxane binding pocket. The overall Cut7 footprint on the S. pombe microtubule surface is altered compared to mammalian tubulin microtubules because of their different polymer architectures. However, the core motor-microtubule interaction is tightly conserved, reflected in similar Cut7 ATPase activities on each microtubule type. AMPPNP-bound Cut7 adopts a kinesin-conserved ATP-like conformation including cover neck bundle formation. However, the Cut7 ATPase is not blocked by a mammalian-specific kinesin-5 inhibitor, consistent with the non-conserved sequence and structure of its loop5 insertion.
Keywords: 3D reconstruction; Cut7; cytoskeleton; mitosis; motor.
Copyright © 2019 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


Similar articles
-
Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism.Proc Natl Acad Sci U S A. 2016 Nov 22;113(47):E7483-E7489. doi: 10.1073/pnas.1611581113. Epub 2016 Nov 9. Proc Natl Acad Sci U S A. 2016. PMID: 27834216 Free PMC article.
-
Protein complexes at the microtubule organizing center regulate bipolar spindle assembly.Cell Cycle. 2008 May 1;7(9):1246-53. doi: 10.4161/cc.7.9.5808. Epub 2008 Feb 22. Cell Cycle. 2008. PMID: 18418055
-
Suppressor Analysis Uncovers That MAPs and Microtubule Dynamics Balance with the Cut7/Kinesin-5 Motor for Mitotic Spindle Assembly in Schizosaccharomyces pombe.G3 (Bethesda). 2019 Jan 9;9(1):269-280. doi: 10.1534/g3.118.200896. G3 (Bethesda). 2019. PMID: 30463883 Free PMC article.
-
Interaction of kinesin motors, microtubules, and MAPs.J Muscle Res Cell Motil. 2006;27(2):125-37. doi: 10.1007/s10974-005-9051-4. Epub 2005 Dec 17. J Muscle Res Cell Motil. 2006. PMID: 16362723 Review.
-
Prime movers: the mechanochemistry of mitotic kinesins.Nat Rev Mol Cell Biol. 2014 Apr;15(4):257-71. doi: 10.1038/nrm3768. Nat Rev Mol Cell Biol. 2014. PMID: 24651543 Review.
Cited by
-
Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.J Biol Chem. 2021 Nov;297(5):101063. doi: 10.1016/j.jbc.2021.101063. Epub 2021 Aug 8. J Biol Chem. 2021. PMID: 34375637 Free PMC article.
-
Distinct regions of the kinesin-5 C-terminal tail are essential for mitotic spindle midzone localization and sliding force.Proc Natl Acad Sci U S A. 2023 Sep 26;120(39):e2306480120. doi: 10.1073/pnas.2306480120. Epub 2023 Sep 19. Proc Natl Acad Sci U S A. 2023. PMID: 37725645 Free PMC article.
-
A Comprehensive Study on the Electrostatic Properties of Tubulin-Tubulin Complexes in Microtubules.Cells. 2023 Jan 5;12(2):238. doi: 10.3390/cells12020238. Cells. 2023. PMID: 36672172 Free PMC article.
-
The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding.Elife. 2020 Jan 20;9:e51131. doi: 10.7554/eLife.51131. Elife. 2020. PMID: 31958056 Free PMC article.
-
A kinesin-1 variant reveals motor-induced microtubule damage in cells.Curr Biol. 2022 Jun 6;32(11):2416-2429.e6. doi: 10.1016/j.cub.2022.04.020. Epub 2022 May 2. Curr Biol. 2022. PMID: 35504282 Free PMC article.
References
-
- Goulet A., Moores C. New insights into the mechanism of force generation by kinesin-5 molecular motors. Int. Rev. Cell Mol. Biol. 2013;304:419–466. - PubMed
-
- Hagan I., Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7 + gene. Nature. 1990;347:563–566. - PubMed
-
- Kapitein L.C., Peterman E.J., Kwok B.H., Kim J.H., Kapoor T.M., Schmidt C.F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. - PubMed
-
- Roostalu J., Hentrich C., Bieling P., Telley I.A., Schiebel E., Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

