Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly
- PMID: 30622273
- PMCID: PMC6325239
- DOI: 10.1038/s41467-018-08028-6
Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly
Abstract
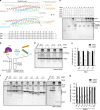
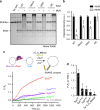
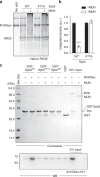
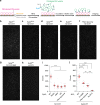
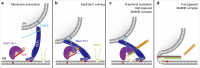
The transition of the Munc18-1/syntaxin-1 complex to the SNARE complex, a key step involved in exocytosis, is regulated by Munc13-1, SNAP-25 and synaptobrevin-2, but the underlying mechanism remains elusive. Here, we identify an interaction between Munc13-1 and the membrane-proximal linker region of synaptobrevin-2, and reveal its essential role in transition and exocytosis. Upon this interaction, Munc13-1 not only recruits synaptobrevin-2-embedded vesicles to the target membrane but also renders the synaptobrevin-2 SNARE motif more accessible to the Munc18-1/syntaxin-1 complex. Afterward, the entry of SNAP-25 leads to a half-zippered SNARE assembly, which eventually dissociates the Munc18-1/syntaxin-1 complex to complete SNARE complex formation. Our data suggest that Munc18-1 and Munc13-1 together serve as a functional template to orchestrate SNARE complex assembly.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Munc13-1 MUN domain and Munc18-1 cooperatively chaperone SNARE assembly through a tetrameric complex.Proc Natl Acad Sci U S A. 2020 Jan 14;117(2):1036-1041. doi: 10.1073/pnas.1914361117. Epub 2019 Dec 30. Proc Natl Acad Sci U S A. 2020. PMID: 31888993 Free PMC article.
-
Tomosyn guides SNARE complex formation in coordination with Munc18 and Munc13.FEBS Lett. 2018 Apr;592(7):1161-1172. doi: 10.1002/1873-3468.13018. Epub 2018 Mar 13. FEBS Lett. 2018. PMID: 29485200
-
Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly.EMBO J. 2020 Aug 17;39(16):e103631. doi: 10.15252/embj.2019103631. Epub 2020 Jul 9. EMBO J. 2020. PMID: 32643828 Free PMC article.
-
Neuronal SNARE complex assembly guided by Munc18-1 and Munc13-1.FEBS Open Bio. 2022 Nov;12(11):1939-1957. doi: 10.1002/2211-5463.13394. Epub 2022 Mar 22. FEBS Open Bio. 2022. PMID: 35278279 Free PMC article. Review.
-
Molecular Mechanisms Underlying Neurotransmitter Release.Annu Rev Biophys. 2022 May 9;51:377-408. doi: 10.1146/annurev-biophys-111821-104732. Epub 2022 Feb 15. Annu Rev Biophys. 2022. PMID: 35167762 Free PMC article. Review.
Cited by
-
Functional role of UNC13D in immune diseases and its therapeutic applications.Front Immunol. 2024 Oct 14;15:1460882. doi: 10.3389/fimmu.2024.1460882. eCollection 2024. Front Immunol. 2024. PMID: 39469717 Free PMC article. Review.
-
K+ Channel-SEC11 Binding Exchange Regulates SNARE Assembly for Secretory Traffic.Plant Physiol. 2019 Nov;181(3):1096-1113. doi: 10.1104/pp.19.00919. Epub 2019 Sep 23. Plant Physiol. 2019. PMID: 31548266 Free PMC article.
-
Munc13-1 MUN domain and Munc18-1 cooperatively chaperone SNARE assembly through a tetrameric complex.Proc Natl Acad Sci U S A. 2020 Jan 14;117(2):1036-1041. doi: 10.1073/pnas.1914361117. Epub 2019 Dec 30. Proc Natl Acad Sci U S A. 2020. PMID: 31888993 Free PMC article.
-
(M)Unc13s in Active Zone Diversity: A Drosophila Perspective.Front Synaptic Neurosci. 2022 Jan 3;13:798204. doi: 10.3389/fnsyn.2021.798204. eCollection 2021. Front Synaptic Neurosci. 2022. PMID: 35046788 Free PMC article. Review.
-
Structural Roles for the Juxtamembrane Linker Region and Transmembrane Region of Synaptobrevin 2 in Membrane Fusion.Front Cell Dev Biol. 2021 Jan 6;8:609708. doi: 10.3389/fcell.2020.609708. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33490074 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

