Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing
- PMID: 30606613
- PMCID: PMC6336504
- DOI: 10.1016/j.neuron.2018.12.006
Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing
Abstract
Microglia are increasingly recognized for their major contributions during brain development and neurodegenerative disease. It is currently unknown whether these functions are carried out by subsets of microglia during different stages of development and adulthood or within specific brain regions. Here, we performed deep single-cell RNA sequencing (scRNA-seq) of microglia and related myeloid cells sorted from various regions of embryonic, early postnatal, and adult mouse brains. We found that the majority of adult microglia expressing homeostatic genes are remarkably similar in transcriptomes, regardless of brain region. By contrast, early postnatal microglia are more heterogeneous. We discovered a proliferative-region-associated microglia (PAM) subset, mainly found in developing white matter, that shares a characteristic gene signature with degenerative disease-associated microglia (DAM). Such PAM have amoeboid morphology, are metabolically active, and phagocytose newly formed oligodendrocytes. This scRNA-seq atlas will be a valuable resource for dissecting innate immune functions in health and disease.
Keywords: brain myeloid cells; brain regions; cell cycle; development; disease-associated microglia; heterogeneity; microglia; phagocytosis; proliferative-region-associated microglia; single-cell RNA-seq.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures






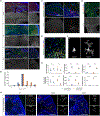
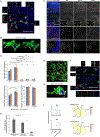
Comment in
-
Demystifying Microglia: And Now the Work Begins.Immunity. 2019 Jan 15;50(1):11-13. doi: 10.1016/j.immuni.2018.12.025. Immunity. 2019. PMID: 30650371
Similar articles
-
Unique microglia expression profile in developing white matter.BMC Res Notes. 2019 Jul 1;12(1):367. doi: 10.1186/s13104-019-4410-1. BMC Res Notes. 2019. PMID: 31262353 Free PMC article.
-
Unique transcriptome signature of mouse microglia.Glia. 2013 Sep;61(9):1429-42. doi: 10.1002/glia.22524. Epub 2013 Jul 8. Glia. 2013. PMID: 23832717
-
RNA sequencing analysis reveals quiescent microglia isolation methods from postnatal mouse brains and limitations of BV2 cells.J Neuroinflammation. 2018 May 22;15(1):153. doi: 10.1186/s12974-018-1195-4. J Neuroinflammation. 2018. PMID: 29788964 Free PMC article.
-
Microglia Function in the Normal Brain.Adv Exp Med Biol. 2016;949:67-92. doi: 10.1007/978-3-319-40764-7_4. Adv Exp Med Biol. 2016. PMID: 27714685 Review.
-
The Myeloid Cell Compartment-Cell by Cell.Annu Rev Immunol. 2019 Apr 26;37:269-293. doi: 10.1146/annurev-immunol-042718-041728. Epub 2019 Jan 16. Annu Rev Immunol. 2019. PMID: 30649988 Review.
Cited by
-
Microglia control small vessel calcification via TREM2.Sci Adv. 2021 Feb 26;7(9):eabc4898. doi: 10.1126/sciadv.abc4898. Print 2021 Feb. Sci Adv. 2021. PMID: 33637522 Free PMC article.
-
Immunometabolism of Tissue-Resident Macrophages - An Appraisal of the Current Knowledge and Cutting-Edge Methods and Technologies.Front Immunol. 2021 May 7;12:665782. doi: 10.3389/fimmu.2021.665782. eCollection 2021. Front Immunol. 2021. PMID: 34025667 Free PMC article. Review.
-
Towards a definition of microglia heterogeneity.Commun Biol. 2022 Oct 20;5(1):1114. doi: 10.1038/s42003-022-04081-6. Commun Biol. 2022. PMID: 36266565 Free PMC article. Review.
-
Single-cell RNA transcriptome analysis of CNS immune cells reveals CXCL16/CXCR6 as maintenance factors for tissue-resident T cells that drive synapse elimination.Genome Med. 2022 Sep 24;14(1):108. doi: 10.1186/s13073-022-01111-0. Genome Med. 2022. PMID: 36153630 Free PMC article.
-
Single-Cell Transcriptomics and In Situ Morphological Analyses Reveal Microglia Heterogeneity Across the Nigrostriatal Pathway.Front Immunol. 2021 Mar 29;12:639613. doi: 10.3389/fimmu.2021.639613. eCollection 2021. Front Immunol. 2021. PMID: 33854507 Free PMC article.
References
-
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, and Raff MC (1992). Cell death and control of cell survival in the oligodendrocyte lineage. Cell 70, 31–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

