Regulatory functions of the Mediator kinases CDK8 and CDK19
- PMID: 30585107
- PMCID: PMC6602567
- DOI: 10.1080/21541264.2018.1556915
Regulatory functions of the Mediator kinases CDK8 and CDK19
Abstract
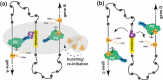
The Mediator-associated kinases CDK8 and CDK19 function in the context of three additional proteins: CCNC and MED12, which activate CDK8/CDK19 kinase function, and MED13, which enables their association with the Mediator complex. The Mediator kinases affect RNA polymerase II (pol II) transcription indirectly, through phosphorylation of transcription factors and by controlling Mediator structure and function. In this review, we discuss cellular roles of the Mediator kinases and mechanisms that enable their biological functions. We focus on sequence-specific, DNA-binding transcription factors and other Mediator kinase substrates, and how CDK8 or CDK19 may enable metabolic and transcriptional reprogramming through enhancers and chromatin looping. We also summarize Mediator kinase inhibitors and their therapeutic potential. Throughout, we note conserved and divergent functions between yeast and mammalian CDK8, and highlight many aspects of kinase module function that remain enigmatic, ranging from potential roles in pol II promoter-proximal pausing to liquid-liquid phase separation.
Keywords: Mediator kinase; RNA polymerase II; chromatin; enhancer; transcription.
Figures
Similar articles
-
Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19.J Biol Chem. 2018 Mar 30;293(13):4870-4882. doi: 10.1074/jbc.RA118.001725. Epub 2018 Feb 13. J Biol Chem. 2018. PMID: 29440396 Free PMC article.
-
The Mediator kinase module: an interface between cell signaling and transcription.Trends Biochem Sci. 2022 Apr;47(4):314-327. doi: 10.1016/j.tibs.2022.01.002. Epub 2022 Feb 19. Trends Biochem Sci. 2022. PMID: 35193797 Free PMC article. Review.
-
Identification of target genes for the CDK subunits of the Mediator complex.Genes Cells. 2011 Dec;16(12):1208-18. doi: 10.1111/j.1365-2443.2011.01565.x. Genes Cells. 2011. PMID: 22117896
-
Mediator kinase module proteins, genetic alterations and expression of super-enhancer regulated genes in colorectal cancer.Pharmacol Rep. 2024 Jun;76(3):535-556. doi: 10.1007/s43440-024-00589-2. Epub 2024 Apr 11. Pharmacol Rep. 2024. PMID: 38602606
-
Expression of CDK8 and CDK8-interacting Genes as Potential Biomarkers in Breast Cancer.Curr Cancer Drug Targets. 2015;15(8):739-49. doi: 10.2174/156800961508151001105814. Curr Cancer Drug Targets. 2015. PMID: 26452386 Free PMC article. Review.
Cited by
-
Transcriptional Antagonism by CDK8 Inhibition Improves Therapeutic Efficacy of MEK Inhibitors.Cancer Res. 2023 Jan 18;83(2):285-300. doi: 10.1158/0008-5472.CAN-21-4309. Cancer Res. 2023. PMID: 36398965 Free PMC article.
-
The Cyclin-Dependent Kinase 8 (CDK8) Inhibitor DCA Promotes a Tolerogenic Chemical Immunophenotype in CD4+ T Cells via a Novel CDK8-GATA3-FOXP3 Pathway.Mol Cell Biol. 2021 Aug 24;41(9):e0008521. doi: 10.1128/MCB.00085-21. Epub 2021 Aug 24. Mol Cell Biol. 2021. PMID: 34124936 Free PMC article.
-
XLID syndrome gene Med12 promotes Ig isotype switching through chromatin modification and enhancer RNA regulation.Sci Adv. 2022 Nov 25;8(47):eadd1466. doi: 10.1126/sciadv.add1466. Epub 2022 Nov 25. Sci Adv. 2022. PMID: 36427307 Free PMC article.
-
Predicting protein and pathway associations for understudied dark kinases using pattern-constrained knowledge graph embedding.PeerJ. 2023 Oct 18;11:e15815. doi: 10.7717/peerj.15815. eCollection 2023. PeerJ. 2023. PMID: 37868056 Free PMC article.
-
DNA Damage, n-3 Long-Chain PUFA Levels and Proteomic Profile in Brazilian Children and Adolescents.Nutrients. 2021 Jul 21;13(8):2483. doi: 10.3390/nu13082483. Nutrients. 2021. PMID: 34444642 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
