7-Methylguanosine Modifications in Transfer RNA (tRNA)
- PMID: 30562954
- PMCID: PMC6320965
- DOI: 10.3390/ijms19124080
7-Methylguanosine Modifications in Transfer RNA (tRNA)
Abstract
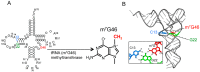
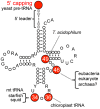
More than 90 different modified nucleosides have been identified in tRNA. Among the tRNA modifications, the 7-methylguanosine (m⁷G) modification is found widely in eubacteria, eukaryotes, and a few archaea. In most cases, the m⁷G modification occurs at position 46 in the variable region and is a product of tRNA (m⁷G46) methyltransferase. The m⁷G46 modification forms a tertiary base pair with C13-G22, and stabilizes the tRNA structure. A reaction mechanism for eubacterial tRNA m⁷G methyltransferase has been proposed based on the results of biochemical, bioinformatic, and structural studies. However, an experimentally determined mechanism of methyl-transfer remains to be ascertained. The physiological functions of m⁷G46 in tRNA have started to be determined over the past decade. For example, tRNA m⁷G46 or tRNA (m⁷G46) methyltransferase controls the amount of other tRNA modifications in thermophilic bacteria, contributes to the pathogenic infectivity, and is also associated with several diseases. In this review, information of tRNA m⁷G modifications and tRNA m⁷G methyltransferases is summarized and the differences in reaction mechanism between tRNA m⁷G methyltransferase and rRNA or mRNA m⁷G methylation enzyme are discussed.
Keywords: RNA modification; methylase; tRNA methyltransferase; tRNA modification.
Conflict of interest statement
The author declares no conflict of interest.
Figures

Similar articles
-
Substrate tRNA recognition mechanism of eubacterial tRNA (m1A58) methyltransferase (TrmI).J Biol Chem. 2015 Feb 27;290(9):5912-25. doi: 10.1074/jbc.M114.606038. Epub 2015 Jan 15. J Biol Chem. 2015. PMID: 25593312 Free PMC article.
-
Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA.RNA. 2002 Oct;8(10):1253-66. doi: 10.1017/s1355838202024019. RNA. 2002. PMID: 12403464 Free PMC article.
-
Structural requirements for the formation of 1-methylguanosine in vivo in tRNA(Pro)GGG of Salmonella typhimurium.J Mol Biol. 1997 Feb 21;266(2):283-96. doi: 10.1006/jmbi.1996.0789. J Mol Biol. 1997. PMID: 9047363
-
tRNA-m1G methyltransferase interactions: touching bases with structure.Biochimie. 1995;77(1-2):62-5. doi: 10.1016/0300-9084(96)88105-3. Biochimie. 1995. PMID: 7599277 Review.
-
Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA.Biomolecules. 2017 Feb 28;7(1):23. doi: 10.3390/biom7010023. Biomolecules. 2017. PMID: 28264529 Free PMC article. Review.
Cited by
-
m7G regulator-mediated methylation modification patterns define immune cell infiltration and patient survival.Front Immunol. 2022 Oct 28;13:1022720. doi: 10.3389/fimmu.2022.1022720. eCollection 2022. Front Immunol. 2022. PMID: 36389726 Free PMC article.
-
N7-methylguanosine-related lncRNAs: Distinction between hot and cold tumors and construction of predictive models in colon adenocarcinoma.Front Oncol. 2022 Sep 15;12:951452. doi: 10.3389/fonc.2022.951452. eCollection 2022. Front Oncol. 2022. PMID: 36185235 Free PMC article.
-
Writers, readers, and erasers RNA modifications and drug resistance in cancer.Mol Cancer. 2024 Aug 30;23(1):178. doi: 10.1186/s12943-024-02089-6. Mol Cancer. 2024. PMID: 39215288 Free PMC article. Review.
-
A Benzophenone-Based Photocaging Strategy for the N7 Position of Guanosine.Angew Chem Int Ed Engl. 2020 Feb 17;59(8):3161-3165. doi: 10.1002/anie.201914573. Epub 2019 Dec 30. Angew Chem Int Ed Engl. 2020. PMID: 31747109 Free PMC article.
-
Analysis of m7G methylation modification patterns and pulmonary vascular immune microenvironment in pulmonary arterial hypertension.Front Immunol. 2022 Dec 5;13:1014509. doi: 10.3389/fimmu.2022.1014509. eCollection 2022. Front Immunol. 2022. PMID: 36544768 Free PMC article.
References
-
- Boccaletto P., Machnicka M.A., Purta E., Pi Atkowski P.-L., Zej Bagí Nski B.-L., Wirecki T.K., De Crécy V., Crécy-Lagard C., Ross R., Limbach P.A., et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2017;46:303–307. doi: 10.1093/nar/gkx1030. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

