Identification of a First Human Norovirus CD8+ T Cell Epitope Restricted to HLA-A*0201 Allele
- PMID: 30542352
- PMCID: PMC6277766
- DOI: 10.3389/fimmu.2018.02782
Identification of a First Human Norovirus CD8+ T Cell Epitope Restricted to HLA-A*0201 Allele
Abstract
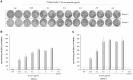
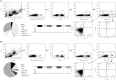
Norovirus (NoV) causes a substantial global burden of acute gastroenteritis in all age groups and the development of NoV vaccine is a high priority. There are still gaps in understanding of protective NoV-specific immunity. Antibody mediated immune responses have been widely studied, but in contrast, the research on NoV-specific human T cell-mediated immunity is very limited. We have recently reported NoV capsid VP1-specific 18-mer peptide (134SPSQVTMFPHIIVDVRQL151) to induce strong CD8+ T cell immune responses in healthy adult donors. This work extends to identify the precise NoV T cell epitope and the restricting human leucocyte antigen (HLA). Pentamer technology was used to detect HLA-A*0201-restricted T cell-mediated responses to 10-mer peptide 139TMFPHIIVDV148 of four healthy adult blood donors. Immunogenicity of the 10-mer epitope was confirmed by ELISPOT IFN-γ and intracellular cytokine staining (ICS) on flow cytometry. A population of CD3+CD8+ T lymphocytes binding to HLA-A*0201/TMFPHIIVDV pentamers was identified in two HLA-A*0201-positive donors. Recognition of the 10-mer epitope by T cells resulted in a strong IFN-γ secretion as shown by ELISPOT assay. In addition, ICS confirmed that high proportion (31 and 59%) of the TMFPHIIVDV epitope-responsive CD3+CD8+ T cells in the two donors had multifunctional phenotype, simultaneously producing IFN-γ, IL-2 and TNF-α cytokines. In the present study novel human NoV HLA-A*0201-restricted minimal 10-mer epitope 139TMFPHIIVDV148 in the capsid VP1 was identified. The HLA-peptide pentamer staining of T cells from healthy donor PBMCs and cytokine responses in ex-vivo ELISPOT and ICS assays suggest that this epitope is recognized during NoV infection and activates memory phenotype of the epitope-specific multifunctional CD8+ T cells. The importance of this epitope in protection from NoV infection remains to be determined.
Keywords: CD8 T cell epitope; ELISPOT IFN-gamma; HLA-A2*0201; cellular immunity; multifunctional T cells; norovirus; pentamer.
Figures


Similar articles
-
The Antigenic Topology of Norovirus as Defined by B and T Cell Epitope Mapping: Implications for Universal Vaccines and Therapeutics.Viruses. 2019 May 10;11(5):432. doi: 10.3390/v11050432. Viruses. 2019. PMID: 31083353 Free PMC article. Review.
-
Specificity of T cells in synovial fluid: high frequencies of CD8(+) T cells that are specific for certain viral epitopes.Arthritis Res. 2000;2(2):154-64. doi: 10.1186/ar80. Epub 2000 Feb 7. Arthritis Res. 2000. PMID: 11062606 Free PMC article.
-
Design of multi-epitope peptides containing HLA class-I and class-II-restricted epitopes derived from immunogenic Leishmania proteins, and evaluation of CD4+ and CD8+ T cell responses induced in cured cutaneous leishmaniasis subjects.PLoS Negl Trop Dis. 2020 Mar 16;14(3):e0008093. doi: 10.1371/journal.pntd.0008093. eCollection 2020 Mar. PLoS Negl Trop Dis. 2020. PMID: 32176691 Free PMC article.
-
Norovirus-Specific CD8+ T Cell Responses in Human Blood and Tissues.Cell Mol Gastroenterol Hepatol. 2021;11(5):1267-1289. doi: 10.1016/j.jcmgh.2020.12.012. Epub 2021 Jan 11. Cell Mol Gastroenterol Hepatol. 2021. PMID: 33444817 Free PMC article.
-
Advances in the study of HLA-restricted epitope vaccines.Hum Vaccin Immunother. 2013 Dec;9(12):2566-77. doi: 10.4161/hv.26088. Epub 2013 Aug 16. Hum Vaccin Immunother. 2013. PMID: 23955319 Free PMC article. Review.
Cited by
-
Virus-Host Interactions Between Nonsecretors and Human Norovirus.Cell Mol Gastroenterol Hepatol. 2020;10(2):245-267. doi: 10.1016/j.jcmgh.2020.03.006. Epub 2020 Apr 11. Cell Mol Gastroenterol Hepatol. 2020. PMID: 32289501 Free PMC article.
-
The Antigenic Topology of Norovirus as Defined by B and T Cell Epitope Mapping: Implications for Universal Vaccines and Therapeutics.Viruses. 2019 May 10;11(5):432. doi: 10.3390/v11050432. Viruses. 2019. PMID: 31083353 Free PMC article. Review.
-
Characteristics of Norovirus capsid protein-specific CD8+ T-Cell responses in previously infected individuals.Virulence. 2024 Dec;15(1):2360133. doi: 10.1080/21505594.2024.2360133. Epub 2024 May 29. Virulence. 2024. PMID: 38803081 Free PMC article.
-
Generation of Norovirus-Specific T Cells From Human Donors With Extensive Cross-Reactivity to Variant Sequences: Implications for Immunotherapy.J Infect Dis. 2020 Feb 3;221(4):578-588. doi: 10.1093/infdis/jiz491. J Infect Dis. 2020. PMID: 31562500 Free PMC article.
-
Immune responses in healthy adults elicited by a bivalent norovirus vaccine candidate composed of GI.4 and GII.4 VLPs without adjuvant.Front Immunol. 2023 Jun 26;14:1188431. doi: 10.3389/fimmu.2023.1188431. eCollection 2023. Front Immunol. 2023. PMID: 37435073 Free PMC article. Clinical Trial.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

