ROBO2 is a stroma suppressor gene in the pancreas and acts via TGF-β signalling
- PMID: 30504844
- PMCID: PMC6269509
- DOI: 10.1038/s41467-018-07497-z
ROBO2 is a stroma suppressor gene in the pancreas and acts via TGF-β signalling
Abstract
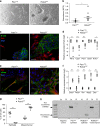
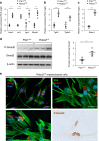
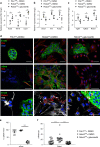
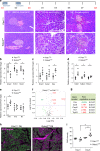
Whereas genomic aberrations in the SLIT-ROBO pathway are frequent in pancreatic ductal adenocarcinoma (PDAC), their function in the pancreas is unclear. Here we report that in pancreatitis and PDAC mouse models, epithelial Robo2 expression is lost while Robo1 expression becomes most prominent in the stroma. Cell cultures of mice with loss of epithelial Robo2 (Pdx1Cre;Robo2F/F) show increased activation of Robo1+ myofibroblasts and induction of TGF-β and Wnt pathways. During pancreatitis, Pdx1Cre;Robo2F/F mice present enhanced myofibroblast activation, collagen crosslinking, T-cell infiltration and tumorigenic immune markers. The TGF-β inhibitor galunisertib suppresses these effects. In PDAC patients, ROBO2 expression is overall low while ROBO1 is variably expressed in epithelium and high in stroma. ROBO2low;ROBO1high patients present the poorest survival. In conclusion, Robo2 acts non-autonomously as a stroma suppressor gene by restraining myofibroblast activation and T-cell infiltration. ROBO1/2 expression in PDAC patients may guide therapy with TGF-β inhibitors or other stroma /immune modulating agents.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Mouse Models Shed Light on the SLIT/ROBO Pathway in Pancreatic Development and Cancer.Trends Cancer. 2019 Mar;5(3):145-148. doi: 10.1016/j.trecan.2019.02.004. Epub 2019 Feb 16. Trends Cancer. 2019. PMID: 30898261
Similar articles
-
Disrupted Slit-Robo signalling results in membranous ventricular septum defects and bicuspid aortic valves.Cardiovasc Res. 2015 Apr 1;106(1):55-66. doi: 10.1093/cvr/cvv040. Epub 2015 Feb 17. Cardiovasc Res. 2015. PMID: 25691540 Free PMC article.
-
Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling.Int J Cancer. 2014 Sep 1;135(5):1110-8. doi: 10.1002/ijc.28765. Epub 2014 May 9. Int J Cancer. 2014. PMID: 24500968
-
Crucial roles of Robo proteins in midline crossing of cerebellofugal axons and lack of their up-regulation after midline crossing.Neural Dev. 2008 Nov 5;3:29. doi: 10.1186/1749-8104-3-29. Neural Dev. 2008. PMID: 18986510 Free PMC article.
-
TGF-β Inhibitors in Metastatic Pancreatic Ductal Adenocarcinoma.J Gastrointest Cancer. 2019 Jun;50(2):207-213. doi: 10.1007/s12029-018-00195-5. J Gastrointest Cancer. 2019. PMID: 30891677 Review.
-
Molecular mechanisms of pancreatic myofibroblast activation in chronic pancreatitis and pancreatic ductal adenocarcinoma.J Gastroenterol. 2021 Aug;56(8):689-703. doi: 10.1007/s00535-021-01800-4. Epub 2021 Jul 19. J Gastroenterol. 2021. PMID: 34279724 Free PMC article. Review.
Cited by
-
Tumour-reprogrammed stromal BCAT1 fuels branched-chain ketoacid dependency in stromal-rich PDAC tumours.Nat Metab. 2020 Aug;2(8):775-792. doi: 10.1038/s42255-020-0226-5. Epub 2020 Jul 6. Nat Metab. 2020. PMID: 32694827 Free PMC article.
-
Tumor-Derived Exosomal miR-29b Reduces Angiogenesis in Pancreatic Cancer by Silencing ROBO1 and SRGAP2.J Immunol Res. 2022 Oct 14;2022:4769385. doi: 10.1155/2022/4769385. eCollection 2022. J Immunol Res. 2022. PMID: 36277474 Free PMC article.
-
Downregulation of Roundabout guidance receptor 2 suppresses hepatocellular carcinoma progression by interacting with Y-box binding protein 1.Sci Rep. 2024 Jan 31;14(1):2588. doi: 10.1038/s41598-024-53013-3. Sci Rep. 2024. PMID: 38297025 Free PMC article.
-
The role of collagen in cancer: from bench to bedside.J Transl Med. 2019 Sep 14;17(1):309. doi: 10.1186/s12967-019-2058-1. J Transl Med. 2019. PMID: 31521169 Free PMC article. Review.
-
Identification and validation of microRNAs that synergize with miR-34a - a basis for combinatorial microRNA therapeutics.Cell Cycle. 2019 Aug;18(15):1798-1811. doi: 10.1080/15384101.2019.1634956. Epub 2019 Jul 1. Cell Cycle. 2019. PMID: 31258013 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

