ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis
- PMID: 30478421
- PMCID: PMC6614557
- DOI: 10.1038/s41591-018-0241-1
ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis
Abstract
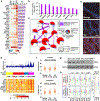
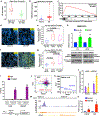
Treatment of prostate cancer (PC) by androgen suppression promotes the emergence of aggressive variants that are androgen receptor (AR) independent. Here we identify the transcription factor ONECUT2 (OC2) as a master regulator of AR networks in metastatic castration-resistant prostate cancer (mCRPC). OC2 acts as a survival factor in mCRPC models, suppresses the AR transcriptional program by direct regulation of AR target genes and the AR licensing factor FOXA1, and activates genes associated with neural differentiation and progression to lethal disease. OC2 appears active in a substantial subset of human prostate adenocarcinoma and neuroendocrine tumors. Inhibition of OC2 by a newly identified small molecule suppresses metastasis in mice. These findings suggest that OC2 displaces AR-dependent growth and survival mechanisms in many cases where AR remains expressed, but where its activity is bypassed. OC2 is also a potential drug target in the metastatic phase of aggressive PC.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
CSMC has pending patent applications PCT/US2017/034768 (Freeman/Rotinen/Murali/You) and US62/548,879 (Freeman/Rotinen/Murali/You) relevant to this study.
Figures




Comment in
-
ONECUT2 many towards AR-independence.Nat Rev Urol. 2019 Feb;16(2):65. doi: 10.1038/s41585-018-0136-4. Nat Rev Urol. 2019. PMID: 30532076 No abstract available.
-
Anticancer drugs: Cutting down on prostate cancer metastases.Nat Rev Drug Discov. 2018 Dec 28;18(1):17. doi: 10.1038/nrd.2018.225. Nat Rev Drug Discov. 2018. PMID: 30591730 No abstract available.
Similar articles
-
ONECUT2 acts as a lineage plasticity driver in adenocarcinoma as well as neuroendocrine variants of prostate cancer.Nucleic Acids Res. 2024 Jul 22;52(13):7740-7760. doi: 10.1093/nar/gkae547. Nucleic Acids Res. 2024. PMID: 38932701 Free PMC article.
-
TCF7 is suppressed by the androgen receptor via microRNA-1-mediated downregulation and is involved in the development of resistance to androgen deprivation in prostate cancer.Prostate Cancer Prostatic Dis. 2017 Jun;20(2):172-178. doi: 10.1038/pcan.2017.2. Epub 2017 Feb 21. Prostate Cancer Prostatic Dis. 2017. PMID: 28220803
-
ONECUT2 is a driver of neuroendocrine prostate cancer.Nat Commun. 2019 Jan 17;10(1):278. doi: 10.1038/s41467-018-08133-6. Nat Commun. 2019. PMID: 30655535 Free PMC article.
-
Emerging Variants of Castration-Resistant Prostate Cancer.Curr Oncol Rep. 2017 May;19(5):32. doi: 10.1007/s11912-017-0593-6. Curr Oncol Rep. 2017. PMID: 28361223 Free PMC article. Review.
-
Shaping Chromatin States in Prostate Cancer by Pioneer Transcription Factors.Cancer Res. 2020 Jun 15;80(12):2427-2436. doi: 10.1158/0008-5472.CAN-19-3447. Epub 2020 Feb 24. Cancer Res. 2020. PMID: 32094298 Free PMC article. Review.
Cited by
-
Meeting report: Metastasis Research Society (MRS) 17th Biennial conference and associated Young Investigator Satellite Meeting (YISM) on cancer metastasis.Clin Exp Metastasis. 2019 Apr;36(2):119-137. doi: 10.1007/s10585-018-09953-y. Epub 2019 Jan 23. Clin Exp Metastasis. 2019. PMID: 30673912
-
Dysregulated Transcriptional Control in Prostate Cancer.Int J Mol Sci. 2019 Jun 13;20(12):2883. doi: 10.3390/ijms20122883. Int J Mol Sci. 2019. PMID: 31200487 Free PMC article. Review.
-
Molecular Links Between Angiogenesis and Neuroendocrine Phenotypes in Prostate Cancer Progression.Front Oncol. 2020 Jan 21;9:1491. doi: 10.3389/fonc.2019.01491. eCollection 2019. Front Oncol. 2020. PMID: 32039001 Free PMC article. Review.
-
It is better to light a candle than to curse the darkness: single-cell transcriptomics sheds new light on pancreas biology and disease.Gut. 2023 Jun;72(6):1211-1219. doi: 10.1136/gutjnl-2022-329313. Epub 2023 Mar 30. Gut. 2023. PMID: 36997301 Free PMC article. Review.
-
Impact of Lineage Plasticity to and from a Neuroendocrine Phenotype on Progression and Response in Prostate and Lung Cancers.Mol Cell. 2020 Nov 19;80(4):562-577. doi: 10.1016/j.molcel.2020.10.033. Mol Cell. 2020. PMID: 33217316 Free PMC article. Review.
References
-
- Sharma NL, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 23, 35–47 (2013). - PubMed
METHODS-ONLY REFERENCES
-
- Bolstad BM, Irizarry RA, Astrand M & Speed TP A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

