Plant HP1 protein ADCP1 links multivalent H3K9 methylation readout to heterochromatin formation
- PMID: 30425322
- PMCID: PMC6318295
- DOI: 10.1038/s41422-018-0104-9
Plant HP1 protein ADCP1 links multivalent H3K9 methylation readout to heterochromatin formation
Abstract
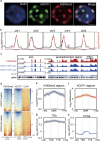
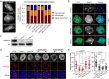
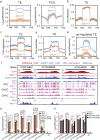
Heterochromatin Protein 1 (HP1) recognizes histone H3 lysine 9 methylation (H3K9me) through its conserved chromodomain and maintains heterochromatin from fission yeast to mammals. However, in Arabidopsis, Like Heterochromatin Protein 1 (LHP1) recognizes and colocalizes genome-wide with H3K27me3, and is the functional homolog of Polycomb protein. This raises the question whether genuine HP1 homologs exist in plants. Here, we report on the discovery of ADCP1, a plant-specific triple tandem Agenet protein, as a multivalent H3K9me reader in Arabidopsis, and establish that ADCP1 is essential for heterochromatin formation and transposon silencing through modulating H3K9 and DNA methylation levels. Structural studies revealed the molecular basis underlying H3K9me-specific recognition by tandem Agenet of ADCP1. Similar to human HP1α and fly HP1a, ADCP1 mediates heterochromatin phase separation. Our results demonstrate that despite its distinct domain compositions, ADCP1 convergently evolves as an HP1-equivalent protein in plants to regulate heterochromatin formation.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
ADCP1: a novel plant H3K9me2 reader.Cell Res. 2019 Jan;29(1):6-7. doi: 10.1038/s41422-018-0119-2. Cell Res. 2019. PMID: 30514899 Free PMC article. No abstract available.
Similar articles
-
Structural basis for the recognition of methylated histone H3 by the Arabidopsis LHP1 chromodomain.J Biol Chem. 2022 Mar;298(3):101623. doi: 10.1016/j.jbc.2022.101623. Epub 2022 Jan 21. J Biol Chem. 2022. PMID: 35074427 Free PMC article.
-
Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly.Mol Cell. 2011 Jan 7;41(1):67-81. doi: 10.1016/j.molcel.2010.12.016. Mol Cell. 2011. PMID: 21211724 Free PMC article.
-
Plant SET- and RING-associated domain proteins in heterochromatinization.Plant J. 2007 Dec;52(5):914-26. doi: 10.1111/j.1365-313X.2007.03286.x. Epub 2007 Sep 22. Plant J. 2007. PMID: 17892444
-
HP1: heterochromatin binding proteins working the genome.Epigenetics. 2010 May 16;5(4):287-92. doi: 10.4161/epi.5.4.11683. Epub 2010 May 3. Epigenetics. 2010. PMID: 20421743 Free PMC article. Review.
-
How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance.Cells. 2020 Jun 12;9(6):1460. doi: 10.3390/cells9061460. Cells. 2020. PMID: 32545538 Free PMC article. Review.
Cited by
-
Get closer and make hotspots: liquid-liquid phase separation in plants.EMBO Rep. 2021 May 5;22(5):e51656. doi: 10.15252/embr.202051656. Epub 2021 Apr 28. EMBO Rep. 2021. PMID: 33913240 Free PMC article. Review.
-
Cis-regulatory sequences in plants: Their importance, discovery, and future challenges.Plant Cell. 2022 Feb 3;34(2):718-741. doi: 10.1093/plcell/koab281. Plant Cell. 2022. PMID: 34918159 Free PMC article. Review.
-
Arabidopsis lamin-like proteins CRWN1 and CRWN2 interact with SUPPRESSOR OF NPR1-1 INDUCIBLE 1 and RAD51D to prevent DNA damage.Plant Cell. 2023 Sep 1;35(9):3345-3362. doi: 10.1093/plcell/koad169. Plant Cell. 2023. PMID: 37335899 Free PMC article.
-
Tidying-up the plant nuclear space: domains, functions, and dynamics.J Exp Bot. 2020 Aug 17;71(17):5160-5178. doi: 10.1093/jxb/eraa282. J Exp Bot. 2020. PMID: 32556244 Free PMC article. Review.
-
Cell type-specific genome scans of DNA methylation divergence indicate an important role for transposable elements.Genome Biol. 2020 Jul 13;21(1):172. doi: 10.1186/s13059-020-02068-2. Genome Biol. 2020. PMID: 32660534 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

