Chemically modified hCFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis
- PMID: 30425265
- PMCID: PMC6233194
- DOI: 10.1038/s41598-018-34960-0
Chemically modified hCFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis
Abstract
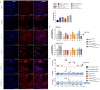
Gene therapy has always been a promising therapeutic approach for Cystic Fibrosis (CF). However, numerous trials using DNA or viral vectors encoding the correct protein resulted in a general low efficacy. In the last years, chemically modified messenger RNA (cmRNA) has been proven to be a highly potent, pulmonary drug. Consequently, we first explored the expression, function and immunogenicity of human (h)CFTR encoded by cmRNAhCFTR in vitro and ex vivo, quantified the expression by flow cytometry, determined its function using a YFP based assay and checked the immune response in human whole blood. Similarly, we examined the function of cmRNAhCFTR in vivo after intratracheal (i.t.) or intravenous (i.v.) injection of the assembled cmRNAhCFTR together with Chitosan-coated PLGA (poly-D, L-lactide-co-glycolide 75:25 (Resomer RG 752 H)) nanoparticles (NPs) by FlexiVent. The amount of expression of human hCFTR encoded by cmRNAhCFTR was quantified by hCFTR ELISA, and cmRNAhCFTR values were assessed by RT-qPCR. Thereby, we observed a significant improvement of lung function, especially in regards to FEV0.1, suggesting NP-cmRNAhCFTR as promising therapeutic option for CF patients independent of their CFTR genotype.
Conflict of interest statement
M.S.D.K. holds a patent on RNA modification (EP2459231B1). M.S.D.K., A.H., A.D. and J.S.A., hold a European patent on delivery of cmRNAh
Figures




Similar articles
-
Optimization of hCFTR lung expression in mice using DNA nanoparticles.Mol Ther. 2012 Jan;20(1):63-72. doi: 10.1038/mt.2011.196. Epub 2011 Sep 27. Mol Ther. 2012. PMID: 21952168 Free PMC article.
-
Human cystic fibrosis transmembrane conductance regulator directed to respiratory epithelial cells of transgenic mice.Nat Genet. 1992 Sep;2(1):13-20. doi: 10.1038/ng0992-13. Nat Genet. 1992. PMID: 1284640
-
Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis.Mol Ther. 2018 Aug 1;26(8):2034-2046. doi: 10.1016/j.ymthe.2018.05.014. Epub 2018 Jun 15. Mol Ther. 2018. PMID: 29910178 Free PMC article.
-
Non-viral approach toward gene therapy of cystic fibrosis lung disease.Curr Gene Ther. 2002 Sep;2(3):295-305. doi: 10.2174/1566523023347832. Curr Gene Ther. 2002. PMID: 12189717 Review.
-
Gene therapy in cystic fibrosis.Chest. 2001 Sep;120(3 Suppl):124S-131S. doi: 10.1378/chest.120.3_suppl.124s. Chest. 2001. PMID: 11555567 Review.
Cited by
-
Unlocking the promise of mRNA therapeutics.Nat Biotechnol. 2022 Nov;40(11):1586-1600. doi: 10.1038/s41587-022-01491-z. Epub 2022 Nov 3. Nat Biotechnol. 2022. PMID: 36329321 Review.
-
Nanomedicine Approaches for the Pulmonary Treatment of Cystic Fibrosis.Front Bioeng Biotechnol. 2019 Dec 17;7:406. doi: 10.3389/fbioe.2019.00406. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31921811 Free PMC article. Review.
-
Cystic Fibrosis: Overview of the Current Development Trends and Innovative Therapeutic Strategies.Pharmaceutics. 2020 Jul 2;12(7):616. doi: 10.3390/pharmaceutics12070616. Pharmaceutics. 2020. PMID: 32630625 Free PMC article. Review.
-
Inhaled RNA Therapy: From Promise to Reality.Trends Pharmacol Sci. 2020 Oct;41(10):715-729. doi: 10.1016/j.tips.2020.08.002. Epub 2020 Sep 4. Trends Pharmacol Sci. 2020. PMID: 32893004 Free PMC article. Review.
-
Empowering gene delivery with protein engineering platforms.Gene Ther. 2023 Dec;30(12):775-782. doi: 10.1038/s41434-022-00379-6. Epub 2022 Dec 19. Gene Ther. 2023. PMID: 36529795 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

