Cryptic binding sites become accessible through surface reconstruction of the type I collagen fibril
- PMID: 30413772
- PMCID: PMC6226522
- DOI: 10.1038/s41598-018-34616-z
Cryptic binding sites become accessible through surface reconstruction of the type I collagen fibril
Abstract
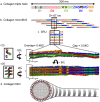
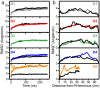
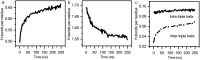
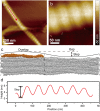
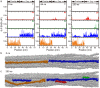
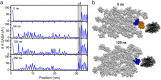
Collagen fibril interactions with cells and macromolecules in the extracellular matrix drive numerous cellular functions. Binding motifs for dozens of collagen-binding proteins have been determined on fully exposed collagen triple helical monomers. However, when the monomers are assembled into the functional collagen fibril, many binding motifs become inaccessible, and yet critical cellular processes occur. Here, we have developed an early stage atomic model of the smallest repeating unit of the type I collagen fibril at the fibril surface that provides a novel framework to address questions about these functionally necessary yet seemingly obstructed interactions. We use an integrative approach by combining molecular dynamics (MD) simulations with atomic force microscopy (AFM) experiments and show that reconstruction of the collagen monomers within the complex fibril play a critical role in collagen interactions. In particular, the fibril surface shows three major conformational changes, which allow cryptic binding sites, including an integrin motif involved in platelet aggregation, to be exposed. The observed dynamics and reconstruction of the fibril surface promote its role as a "smart fibril" to keep certain binding sites cryptic, and to allow accessibility of recognition domains when appropriate.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Revealing Accessibility of Cryptic Protein Binding Sites within the Functional Collagen Fibril.Biomolecules. 2017 Nov 1;7(4):76. doi: 10.3390/biom7040076. Biomolecules. 2017. PMID: 29104255 Free PMC article. Review.
-
Collagen fibril surface displays a constellation of sites capable of promoting fibril assembly, stability, and hemostasis.Connect Tissue Res. 2011 Feb;52(1):18-24. doi: 10.3109/03008207.2010.511354. Epub 2010 Nov 30. Connect Tissue Res. 2011. PMID: 21117898 Free PMC article.
-
Selecting the correct cellular model for assessing of the biological response of collagen-based biomaterials.Acta Biomater. 2018 Jan;65:88-101. doi: 10.1016/j.actbio.2017.10.035. Epub 2017 Oct 26. Acta Biomater. 2018. PMID: 29107054 Free PMC article.
-
Effect of preservation conditions of collagen substrate on its fibril formation and rabbit chondrocyte morphology.J Biosci Bioeng. 2012 Sep;114(3):360-3. doi: 10.1016/j.jbiosc.2012.04.013. Epub 2012 Jun 5. J Biosci Bioeng. 2012. PMID: 22677065
-
Fibrillar Collagens.Subcell Biochem. 2017;82:457-490. doi: 10.1007/978-3-319-49674-0_14. Subcell Biochem. 2017. PMID: 28101870 Review.
Cited by
-
The mechanical cell - the role of force dependencies in synchronising protein interaction networks.J Cell Sci. 2022 Nov 15;135(22):jcs259769. doi: 10.1242/jcs.259769. Epub 2022 Nov 18. J Cell Sci. 2022. PMID: 36398718 Free PMC article. Review.
-
Role of prolyl hydroxylation in the molecular interactions of collagens.Essays Biochem. 2019 Sep 13;63(3):325-335. doi: 10.1042/EBC20180053. Print 2019 Sep 13. Essays Biochem. 2019. PMID: 31350381 Free PMC article. Review.
-
Adhesion force microscopy is sensitive to the charge distribution at the surface of single collagen fibrils.Nanoscale Adv. 2022 Oct 18;4(22):4829-4837. doi: 10.1039/d2na00514j. eCollection 2022 Nov 8. Nanoscale Adv. 2022. PMID: 36381506 Free PMC article.
-
Physiological mechanical forces accelerate the degradation of bovine lung collagen fibers by bacterial collagenase.Sci Rep. 2024 Nov 22;14(1):29008. doi: 10.1038/s41598-024-77704-z. Sci Rep. 2024. PMID: 39578499 Free PMC article.
-
Increased Collagen Turnover Impairs Tendon Microstructure and Stability in Integrin α2β1-Deficient Mice.Int J Mol Sci. 2020 Apr 18;21(8):2835. doi: 10.3390/ijms21082835. Int J Mol Sci. 2020. PMID: 32325713 Free PMC article.
References
-
- Vogel WF. Collagen-receptor signaling in health and disease. Eur J Dermatol. 2001;11:506–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

