Protein Data Bank: the single global archive for 3D macromolecular structure data
- PMID: 30357364
- PMCID: PMC6324056
- DOI: 10.1093/nar/gky949
Protein Data Bank: the single global archive for 3D macromolecular structure data
Abstract
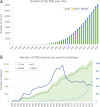
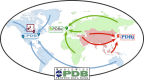
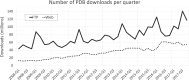
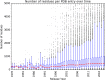
The Protein Data Bank (PDB) is the single global archive of experimentally determined three-dimensional (3D) structure data of biological macromolecules. Since 2003, the PDB has been managed by the Worldwide Protein Data Bank (wwPDB; wwpdb.org), an international consortium that collaboratively oversees deposition, validation, biocuration, and open access dissemination of 3D macromolecular structure data. The PDB Core Archive houses 3D atomic coordinates of more than 144 000 structural models of proteins, DNA/RNA, and their complexes with metals and small molecules and related experimental data and metadata. Structure and experimental data/metadata are also stored in the PDB Core Archive using the readily extensible wwPDB PDBx/mmCIF master data format, which will continue to evolve as data/metadata from new experimental techniques and structure determination methods are incorporated by the wwPDB. Impacts of the recently developed universal wwPDB OneDep deposition/validation/biocuration system and various methods-specific wwPDB Validation Task Forces on improving the quality of structures and data housed in the PDB Core Archive are described together with current challenges and future plans.
Figures
Similar articles
-
The Protein Data Bank Archive.Methods Mol Biol. 2021;2305:3-21. doi: 10.1007/978-1-0716-1406-8_1. Methods Mol Biol. 2021. PMID: 33950382 Review.
-
Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive.Methods Mol Biol. 2017;1607:627-641. doi: 10.1007/978-1-4939-7000-1_26. Methods Mol Biol. 2017. PMID: 28573592 Free PMC article. Review.
-
OneDep: Unified wwPDB System for Deposition, Biocuration, and Validation of Macromolecular Structures in the PDB Archive.Structure. 2017 Mar 7;25(3):536-545. doi: 10.1016/j.str.2017.01.004. Epub 2017 Feb 9. Structure. 2017. PMID: 28190782 Free PMC article.
-
RCSB Protein Data Bank: Sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education.Protein Sci. 2018 Jan;27(1):316-330. doi: 10.1002/pro.3331. Epub 2017 Nov 11. Protein Sci. 2018. PMID: 29067736 Free PMC article. Review.
-
PDBx/mmCIF Ecosystem: Foundational Semantic Tools for Structural Biology.J Mol Biol. 2022 Jun 15;434(11):167599. doi: 10.1016/j.jmb.2022.167599. Epub 2022 Apr 20. J Mol Biol. 2022. PMID: 35460671 Free PMC article. Review.
Cited by
-
CHARMM-GUI PDB Reader and Manipulator: Covalent Ligand Modeling and Simulation.J Mol Biol. 2024 Sep 1;436(17):168554. doi: 10.1016/j.jmb.2024.168554. Epub 2024 Mar 27. J Mol Biol. 2024. PMID: 39237201
-
Inhibitors of Transthyretin Amyloidosis: How to Rank Drug Candidates Using X-ray Crystallography Data.Molecules. 2024 Feb 18;29(4):895. doi: 10.3390/molecules29040895. Molecules. 2024. PMID: 38398647 Free PMC article.
-
EMBL's European Bioinformatics Institute (EMBL-EBI) in 2022.Nucleic Acids Res. 2023 Jan 6;51(D1):D9-D17. doi: 10.1093/nar/gkac1098. Nucleic Acids Res. 2023. PMID: 36477213 Free PMC article.
-
Topological analysis of SARS CoV-2 main protease.Chaos. 2020 Jun;30(6):061102. doi: 10.1063/5.0013029. Chaos. 2020. PMID: 32611087 Free PMC article.
-
Identification of Corosolic and Oleanolic Acids as Molecules Antagonizing the Human RORγT Nuclear Receptor Using the Calculated Fingerprints of the Molecular Similarity.Int J Mol Sci. 2022 Feb 8;23(3):1906. doi: 10.3390/ijms23031906. Int J Mol Sci. 2022. PMID: 35163824 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM109046/GM/NIGMS NIH HHS/United States
- BB/M013146/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 88944/WT_/Wellcome Trust/United Kingdom
- BB/K016970/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M020347/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K020013/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M020428/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011674/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104948/WT_/Wellcome Trust/United Kingdom
- R01 GM133198/GM/NIGMS NIH HHS/United States
- BB/J007471/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/G022577/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

