Defining Essentiality Score of Protein-Coding Genes and Long Noncoding RNAs
- PMID: 30356729
- PMCID: PMC6189311
- DOI: 10.3389/fgene.2018.00380
Defining Essentiality Score of Protein-Coding Genes and Long Noncoding RNAs
Abstract
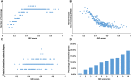
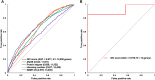
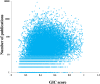
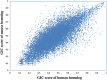
Measuring the essentiality of genes is critically important in biology and medicine. Here we proposed a computational method, GIC (Gene Importance Calculator), which can efficiently predict the essentiality of both protein-coding genes and long noncoding RNAs (lncRNAs) based on only sequence information. For identifying the essentiality of protein-coding genes, GIC outperformed well-established computational scores. In an independent mouse lncRNA dataset, GIC also achieved an exciting performance (AUC = 0.918). In contrast, the traditional computational methods are not applicable to lncRNAs. Moreover, we explored several potential applications of GIC score. Firstly, we revealed a correlation between gene GIC score and research hotspots of genes. Moreover, GIC score can be used to evaluate whether a gene in mouse is representative for its homolog in human by dissecting its cross-species difference. This is critical for basic medicine because many basic medical studies are performed in animal models. Finally, we showed that GIC score can be used to identify candidate genes from a transcriptomics study. GIC is freely available at http://www.cuilab.cn/gic/.
Keywords: essentiality; lncRNAs; machine learning; prediction; protein-coding genes.
Figures

Similar articles
-
miES: predicting the essentiality of miRNAs with machine learning and sequence features.Bioinformatics. 2019 Mar 15;35(6):1053-1054. doi: 10.1093/bioinformatics/bty738. Bioinformatics. 2019. PMID: 30165607
-
LncTar: a tool for predicting the RNA targets of long noncoding RNAs.Brief Bioinform. 2015 Sep;16(5):806-12. doi: 10.1093/bib/bbu048. Epub 2014 Dec 17. Brief Bioinform. 2015. PMID: 25524864
-
CRlncRC: a machine learning-based method for cancer-related long noncoding RNA identification using integrated features.BMC Med Genomics. 2018 Dec 31;11(Suppl 6):120. doi: 10.1186/s12920-018-0436-9. BMC Med Genomics. 2018. PMID: 30598114 Free PMC article.
-
Long non-coding RNAs and complex diseases: from experimental results to computational models.Brief Bioinform. 2017 Jul 1;18(4):558-576. doi: 10.1093/bib/bbw060. Brief Bioinform. 2017. PMID: 27345524 Free PMC article. Review.
-
LncFinder: an integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property.Brief Bioinform. 2019 Nov 27;20(6):2009-2027. doi: 10.1093/bib/bby065. Brief Bioinform. 2019. PMID: 30084867 Free PMC article. Review.
Cited by
-
SGII: Systematic Identification of Essential lncRNAs in Mouse and Human Genome With lncRNA-Protein-Protein Heterogeneous Interaction Network.Front Genet. 2022 Mar 21;13:864564. doi: 10.3389/fgene.2022.864564. eCollection 2022. Front Genet. 2022. PMID: 35386279 Free PMC article.
-
Long Noncoding RNAs in the Pathogenesis of Insulin Resistance.Int J Mol Sci. 2022 Dec 16;23(24):16054. doi: 10.3390/ijms232416054. Int J Mol Sci. 2022. PMID: 36555704 Free PMC article. Review.
-
Importance score of SARS-CoV-2 genome predicts the death risk of COVID-19.Cell Death Discov. 2022 Jul 2;8(1):303. doi: 10.1038/s41420-022-01100-7. Cell Death Discov. 2022. PMID: 35780139 Free PMC article. No abstract available.
-
XGEM: Predicting Essential miRNAs by the Ensembles of Various Sequence-Based Classifiers With XGBoost Algorithm.Front Genet. 2022 Mar 28;13:877409. doi: 10.3389/fgene.2022.877409. eCollection 2022. Front Genet. 2022. PMID: 35419029 Free PMC article.
-
Pig-specific RNA editing during early embryo development revealed by genome-wide comparisons.FEBS Open Bio. 2020 Jul;10(7):1389-1402. doi: 10.1002/2211-5463.12900. Epub 2020 Jun 25. FEBS Open Bio. 2020. PMID: 32433824 Free PMC article.
References
LinkOut - more resources
Full Text Sources

