SUMOgo: Prediction of sumoylation sites on lysines by motif screening models and the effects of various post-translational modifications
- PMID: 30341374
- PMCID: PMC6195521
- DOI: 10.1038/s41598-018-33951-5
SUMOgo: Prediction of sumoylation sites on lysines by motif screening models and the effects of various post-translational modifications
Abstract
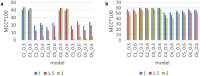
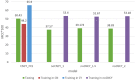
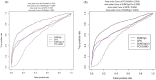
Most modern tools used to predict sites of small ubiquitin-like modifier (SUMO) binding (referred to as SUMOylation) use algorithms, chemical features of the protein, and consensus motifs. However, these tools rarely consider the influence of post-translational modification (PTM) information for other sites within the same protein on the accuracy of prediction results. This study applied the Random Forest machine learning method, as well as motif screening models and a feature selection combination mechanism, to develop a SUMOylation prediction system, referred to as SUMOgo. With regard to prediction method, PTM sites were coded as new functional features in addition to structural features, such as sequence-based binary coding, encoded chemical features of proteins, and encoded secondary structure information that is important for PTM. Twenty cycles of prediction were conducted with a 1:1 combination of positive test data and random negative data. Matthew's correlation coefficient of SUMOgo reached 0.511, which is higher than that of current commonly used tools. This study further verified the important role of PTM in SUMOgo and includes a case study on CREB binding protein (CREBBP). The website for the final tool is http://predictor.nchu.edu.tw/SUMOgo .
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Protein sumoylation sites prediction based on two-stage feature selection.Mol Divers. 2010 Feb;14(1):81-6. doi: 10.1007/s11030-009-9149-5. Epub 2009 May 27. Mol Divers. 2010. PMID: 19472067
-
SUMO-Forest: A Cascade Forest based method for the prediction of SUMOylation sites on imbalanced data.Gene. 2020 May 30;741:144536. doi: 10.1016/j.gene.2020.144536. Epub 2020 Mar 8. Gene. 2020. PMID: 32160959 No abstract available.
-
Predicting protein sumoylation sites from sequence features.Amino Acids. 2012 Jul;43(1):447-55. doi: 10.1007/s00726-011-1100-2. Epub 2011 Oct 7. Amino Acids. 2012. PMID: 21986959
-
Recent Development of Machine Learning Methods in Sumoylation Sites Prediction.Curr Med Chem. 2022;29(5):894-907. doi: 10.2174/0929867328666210915112030. Curr Med Chem. 2022. PMID: 34525906 Review.
-
Large-scale comparative assessment of computational predictors for lysine post-translational modification sites.Brief Bioinform. 2019 Nov 27;20(6):2267-2290. doi: 10.1093/bib/bby089. Brief Bioinform. 2019. PMID: 30285084 Free PMC article. Review.
Cited by
-
Crosstalk between the chloroplast protein import and SUMO systems revealed through genetic and molecular investigation in Arabidopsis.Elife. 2021 Sep 2;10:e60960. doi: 10.7554/eLife.60960. Elife. 2021. PMID: 34473053 Free PMC article.
-
Topotecan and Ginkgolic Acid Inhibit the Expression and Transport Activity of Human Organic Anion Transporter 3 by Suppressing SUMOylation of the Transporter.Pharmaceutics. 2024 May 9;16(5):638. doi: 10.3390/pharmaceutics16050638. Pharmaceutics. 2024. PMID: 38794300 Free PMC article.
-
SUMOylation of α-tubulin is a novel modification regulating microtubule dynamics.J Mol Cell Biol. 2021 May 7;13(2):91-103. doi: 10.1093/jmcb/mjaa076. J Mol Cell Biol. 2021. PMID: 33394042 Free PMC article.
-
SUMOylation controls Hu antigen R posttranscriptional activity in liver cancer.Cell Rep. 2024 Mar 26;43(3):113924. doi: 10.1016/j.celrep.2024.113924. Epub 2024 Mar 18. Cell Rep. 2024. PMID: 38507413 Free PMC article.
-
ADPriboDB v2.0: An Updated Database of ADP-ribosylated Proteins.bioRxiv [Preprint]. 2020 Sep 25:2020.09.24.298851. doi: 10.1101/2020.09.24.298851. bioRxiv. 2020. Update in: Nucleic Acids Res. 2021 Jan 8;49(D1):D261-D265. doi: 10.1093/nar/gkaa941 PMID: 32995784 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

