The architecture of EGFR's basal complexes reveals autoinhibition mechanisms in dimers and oligomers
- PMID: 30337523
- PMCID: PMC6193980
- DOI: 10.1038/s41467-018-06632-0
The architecture of EGFR's basal complexes reveals autoinhibition mechanisms in dimers and oligomers
Abstract
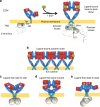
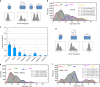
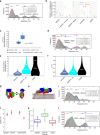
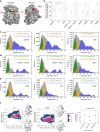
Our current understanding of epidermal growth factor receptor (EGFR) autoinhibition is based on X-ray structural data of monomer and dimer receptor fragments and does not explain how mutations achieve ligand-independent phosphorylation. Using a repertoire of imaging technologies and simulations we reveal an extracellular head-to-head interaction through which ligand-free receptor polymer chains of various lengths assemble. The architecture of the head-to-head interaction prevents kinase-mediated dimerisation. The latter, afforded by mutation or intracellular treatments, splits the autoinhibited head-to-head polymers to form stalk-to-stalk flexible non-extended dimers structurally coupled across the plasma membrane to active asymmetric tyrosine kinase dimers, and extended dimers coupled to inactive symmetric kinase dimers. Contrary to the previously proposed main autoinhibitory function of the inactive symmetric kinase dimer, our data suggest that only dysregulated species bear populations of symmetric and asymmetric kinase dimers that coexist in equilibrium at the plasma membrane under the modulation of the C-terminal domain.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
EGFR oligomerization organizes kinase-active dimers into competent signalling platforms.Nat Commun. 2016 Oct 31;7:13307. doi: 10.1038/ncomms13307. Nat Commun. 2016. PMID: 27796308 Free PMC article.
-
Architecture and membrane interactions of the EGF receptor.Cell. 2013 Jan 31;152(3):557-69. doi: 10.1016/j.cell.2012.12.030. Cell. 2013. PMID: 23374350 Free PMC article.
-
Allosteric regulation of epidermal growth factor (EGF) receptor ligand binding by tyrosine kinase inhibitors.J Biol Chem. 2018 Aug 31;293(35):13401-13414. doi: 10.1074/jbc.RA118.004139. Epub 2018 Jul 11. J Biol Chem. 2018. PMID: 29997256 Free PMC article.
-
Emerging Allosteric Mechanism of EGFR Activation in Physiological and Pathological Contexts.Biophys J. 2019 Jul 9;117(1):5-13. doi: 10.1016/j.bpj.2019.05.021. Epub 2019 May 28. Biophys J. 2019. PMID: 31202480 Free PMC article. Review.
-
Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers.Pharmacol Res. 2019 Jan;139:395-411. doi: 10.1016/j.phrs.2018.11.014. Epub 2018 Nov 27. Pharmacol Res. 2019. PMID: 30500458 Review.
Cited by
-
A perspective of fluorescence microscopy for cellular structural biology with EGFR as witness.J Microsc. 2023 Jul;291(1):73-91. doi: 10.1111/jmi.13151. Epub 2022 Nov 4. J Microsc. 2023. PMID: 36282005 Free PMC article.
-
An Albumin-Holliday Junction Biomolecular Modular Design for Programmable Multifunctionality and Prolonged Circulation.Bioconjug Chem. 2024 Feb 21;35(2):214-222. doi: 10.1021/acs.bioconjchem.3c00491. Epub 2024 Jan 17. Bioconjug Chem. 2024. PMID: 38231391 Free PMC article.
-
Equilibrium Between Dimeric and Monomeric Forms of Human Epidermal Growth Factor is Shifted Towards Dimers in a Solution.Protein J. 2022 Apr;41(2):245-259. doi: 10.1007/s10930-022-10051-y. Epub 2022 Mar 29. Protein J. 2022. PMID: 35348971
-
A recent development of new therapeutic agents and novel drug targets for cancer treatment.SAGE Open Med. 2021 Dec 23;9:20503121211067083. doi: 10.1177/20503121211067083. eCollection 2021. SAGE Open Med. 2021. PMID: 34992782 Free PMC article. Review.
-
Recognition of BRAF by CDC37 and Re-Evaluation of the Activation Mechanism for the Class 2 BRAF-L597R Mutant.Biomolecules. 2022 Jun 28;12(7):905. doi: 10.3390/biom12070905. Biomolecules. 2022. PMID: 35883461 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

