mRNA cap regulation in mammalian cell function and fate
- PMID: 30312682
- PMCID: PMC6414751
- DOI: 10.1016/j.bbagrm.2018.09.011
mRNA cap regulation in mammalian cell function and fate
Abstract
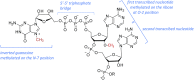
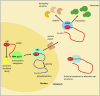
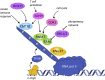
In this review we explore the regulation of mRNA cap formation and its impact on mammalian cells. The mRNA cap is a highly methylated modification of the 5' end of RNA pol II-transcribed RNA. It protects RNA from degradation, recruits complexes involved in RNA processing, export and translation initiation, and marks cellular mRNA as "self" to avoid recognition by the innate immune system. The mRNA cap can be viewed as a unique mark which selects RNA pol II transcripts for specific processing and translation. Over recent years, examples of regulation of mRNA cap formation have emerged, induced by oncogenes, developmental pathways and during the cell cycle. These signalling pathways regulate the rate and extent of mRNA cap formation, resulting in changes in gene expression, cell physiology and cell function.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Regulation of mRNA capping in the cell cycle.RNA Biol. 2017 Jan 2;14(1):11-14. doi: 10.1080/15476286.2016.1251540. Epub 2016 Oct 28. RNA Biol. 2017. PMID: 27791484 Free PMC article. Review.
-
Myc up-regulates formation of the mRNA methyl cap.Biochem Soc Trans. 2010 Dec;38(6):1598-601. doi: 10.1042/BST0381598. Biochem Soc Trans. 2010. PMID: 21118133
-
Enzymology of RNA cap synthesis.Wiley Interdiscip Rev RNA. 2010 Jul-Aug;1(1):152-72. doi: 10.1002/wrna.19. Epub 2010 May 25. Wiley Interdiscip Rev RNA. 2010. PMID: 21956912 Free PMC article. Review.
-
CAPAM: The mRNA Cap Adenosine N6-Methyltransferase.Trends Biochem Sci. 2019 Mar;44(3):183-185. doi: 10.1016/j.tibs.2019.01.002. Epub 2019 Jan 21. Trends Biochem Sci. 2019. PMID: 30679132 Free PMC article.
-
mRNA capping: biological functions and applications.Nucleic Acids Res. 2016 Sep 19;44(16):7511-26. doi: 10.1093/nar/gkw551. Epub 2016 Jun 17. Nucleic Acids Res. 2016. PMID: 27317694 Free PMC article. Review.
Cited by
-
A Comprehensive Review of the Global Efforts on COVID-19 Vaccine Development.ACS Cent Sci. 2021 Apr 28;7(4):512-533. doi: 10.1021/acscentsci.1c00120. Epub 2021 Mar 29. ACS Cent Sci. 2021. PMID: 34056083 Free PMC article.
-
SPAAC-NAD-seq, a sensitive and accurate method to profile NAD+-capped transcripts.Proc Natl Acad Sci U S A. 2021 Mar 30;118(13):e2025595118. doi: 10.1073/pnas.2025595118. Proc Natl Acad Sci U S A. 2021. PMID: 33753511 Free PMC article.
-
mRNA-based therapeutics: powerful and versatile tools to combat diseases.Signal Transduct Target Ther. 2022 May 21;7(1):166. doi: 10.1038/s41392-022-01007-w. Signal Transduct Target Ther. 2022. PMID: 35597779 Free PMC article. Review.
-
Biological roles of RNA m7G modification and its implications in cancer.Biol Direct. 2023 Sep 14;18(1):58. doi: 10.1186/s13062-023-00414-5. Biol Direct. 2023. PMID: 37710294 Free PMC article. Review.
-
Mesenchymal stem cell engineering by ARCA analog-capped mRNA.Mol Ther Nucleic Acids. 2023 Jul 17;33:454-468. doi: 10.1016/j.omtn.2023.07.006. eCollection 2023 Sep 12. Mol Ther Nucleic Acids. 2023. PMID: 37588684 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

