Dengue virus and the host innate immune response
- PMID: 30301880
- PMCID: PMC6177401
- DOI: 10.1038/s41426-018-0168-0
Dengue virus and the host innate immune response
Abstract
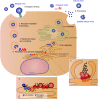
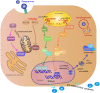
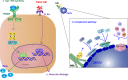
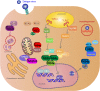
Dengue virus (DENV) is a mosquito-borne Flavivirus that is endemic in many tropical and sub-tropical countries where the transmission vectors Aedes spp. mosquitoes resides. There are four serotypes of the virus. Each serotype is antigenically different, meaning they elicit heterologous antibodies. Infection with one serotype will create neutralizing antibodies to the serotype. Cross-protection from other serotypes is not long term, instead heterotypic infection can cause severe disease. This review will focus on the innate immune response to DENV infection and the virus evasion of the innate immune system by escaping recognition or inhibiting the production of an antiviral state. Activated innate immune pathways includes type I interferon, complement, apoptosis, and autophagy, which the virus can evade or exploit to exacerbate disease. It is important to understand out how the immune system reacts to infection and how the virus evades immune response in order to develop effective antivirals and vaccines.
Figures
Similar articles
-
Innate immunity to dengue virus infection and subversion of antiviral responses.J Mol Biol. 2014 Mar 20;426(6):1148-60. doi: 10.1016/j.jmb.2013.11.023. Epub 2013 Dec 3. J Mol Biol. 2014. PMID: 24316047 Free PMC article. Review.
-
Innate Antiviral Immunity against Dengue Virus.Crit Rev Immunol. 2015;35(3):253-60. doi: 10.1615/critrevimmunol.2015014251. Crit Rev Immunol. 2015. PMID: 26559229 Review.
-
Immune response to dengue virus and prospects for a vaccine.Annu Rev Immunol. 2011;29:587-619. doi: 10.1146/annurev-immunol-031210-101315. Annu Rev Immunol. 2011. PMID: 21219187 Review.
-
Dengue virus replication in infected human keratinocytes leads to activation of antiviral innate immune responses.Infect Genet Evol. 2011 Oct;11(7):1664-73. doi: 10.1016/j.meegid.2011.06.009. Epub 2011 Jun 21. Infect Genet Evol. 2011. PMID: 21722754
-
Evasion of early innate immune response by 2'-O-methylation of dengue genomic RNA.Virology. 2016 Dec;499:259-266. doi: 10.1016/j.virol.2016.09.022. Epub 2016 Oct 4. Virology. 2016. PMID: 27716465 Free PMC article.
Cited by
-
Dengue fever in hyperglycemic patients: an emerging public health concern demanding eyes on the effective management strategies.Health Sci Rep. 2024 Oct 16;7(10):e70144. doi: 10.1002/hsr2.70144. eCollection 2024 Oct. Health Sci Rep. 2024. PMID: 39421212 Free PMC article.
-
Traditional Knowledge to Contemporary Medication in the Treatment of Infectious Disease Dengue: A Review.Front Pharmacol. 2022 Mar 14;13:750494. doi: 10.3389/fphar.2022.750494. eCollection 2022. Front Pharmacol. 2022. PMID: 35359838 Free PMC article. Review.
-
[Baicalin suppresses type 2 dengue virus-induced autophagy of human umbilical vein endothelial cells by inhibiting the PI3K/AKT pathway].Nan Fang Yi Ke Da Xue Xue Bao. 2024 Jul 20;44(7):1272-1283. doi: 10.12122/j.issn.1673-4254.2024.07.07. Nan Fang Yi Ke Da Xue Xue Bao. 2024. PMID: 39051073 Free PMC article. Chinese.
-
Potential Role of Birds in Japanese Encephalitis Virus Zoonotic Transmission and Genotype Shift.Viruses. 2021 Feb 24;13(3):357. doi: 10.3390/v13030357. Viruses. 2021. PMID: 33668224 Free PMC article. Review.
-
Maternal Immunity and Vaccination Influence Disease Severity in Progeny in a Novel Mast Cell-Deficient Mouse Model of Severe Dengue.Viruses. 2021 May 12;13(5):900. doi: 10.3390/v13050900. Viruses. 2021. PMID: 34066286 Free PMC article.
References
-
- Thomas Stephen J., Endy Timothy P., Rothman Alan L. Viral Infections of Humans. Boston, MA: Springer US; 2014. Flaviviruses: Dengue; pp. 351–381.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
