Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris
- PMID: 30283141
- PMCID: PMC6642641
- DOI: 10.1038/s41586-018-0590-4
Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris
Abstract
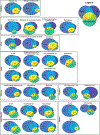
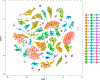
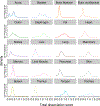
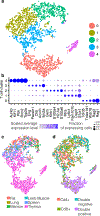
Here we present a compendium of single-cell transcriptomic data from the model organism Mus musculus that comprises more than 100,000 cells from 20 organs and tissues. These data represent a new resource for cell biology, reveal gene expression in poorly characterized cell populations and enable the direct and controlled comparison of gene expression in cell types that are shared between tissues, such as T lymphocytes and endothelial cells from different anatomical locations. Two distinct technical approaches were used for most organs: one approach, microfluidic droplet-based 3'-end counting, enabled the survey of thousands of cells at relatively low coverage, whereas the other, full-length transcript analysis based on fluorescence-activated cell sorting, enabled the characterization of cell types with high sensitivity and coverage. The cumulative data provide the foundation for an atlas of transcriptomic cell biology.
Figures






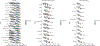







Comment in
-
Navigating mouse cell types.Nat Rev Genet. 2018 Dec;19(12):739. doi: 10.1038/s41576-018-0067-1. Nat Rev Genet. 2018. PMID: 30341441 No abstract available.
-
Cell portrait of a mouse.Nat Methods. 2018 Dec;15(12):1001. doi: 10.1038/s41592-018-0247-0. Nat Methods. 2018. PMID: 30504881 No abstract available.
Similar articles
-
A single-cell transcriptomic atlas characterizes ageing tissues in the mouse.Nature. 2020 Jul;583(7817):590-595. doi: 10.1038/s41586-020-2496-1. Epub 2020 Jul 15. Nature. 2020. PMID: 32669714 Free PMC article.
-
The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans.Science. 2022 May 13;376(6594):eabl4896. doi: 10.1126/science.abl4896. Epub 2022 May 13. Science. 2022. PMID: 35549404 Free PMC article.
-
Ageing hallmarks exhibit organ-specific temporal signatures.Nature. 2020 Jul;583(7817):596-602. doi: 10.1038/s41586-020-2499-y. Epub 2020 Jul 15. Nature. 2020. PMID: 32669715 Free PMC article.
-
Microfluidic single-cell transcriptomics: moving towards multimodal and spatiotemporal omics.Lab Chip. 2021 Oct 12;21(20):3829-3849. doi: 10.1039/d1lc00607j. Lab Chip. 2021. PMID: 34541590 Review.
-
A primer for generating and using transcriptome data and gene sets.Development. 2020 Dec 23;147(24):dev193854. doi: 10.1242/dev.193854. Development. 2020. PMID: 33361089 Free PMC article. Review.
Cited by
-
In-depth analysis reveals complex molecular aetiology in a cohort of idiopathic cerebral palsy.Brain. 2022 Mar 29;145(1):119-141. doi: 10.1093/brain/awab209. Brain. 2022. PMID: 34077496 Free PMC article.
-
Answering open questions in biology using spatial genomics and structured methods.BMC Bioinformatics. 2024 Sep 4;25(1):291. doi: 10.1186/s12859-024-05912-5. BMC Bioinformatics. 2024. PMID: 39232666 Free PMC article. Review.
-
High accuracy gene expression profiling of sorted cell subpopulations from breast cancer PDX model tissue.PLoS One. 2020 Sep 10;15(9):e0238594. doi: 10.1371/journal.pone.0238594. eCollection 2020. PLoS One. 2020. PMID: 32911489 Free PMC article.
-
Capillary cell-type specialization in the alveolus.Nature. 2020 Oct;586(7831):785-789. doi: 10.1038/s41586-020-2822-7. Epub 2020 Oct 14. Nature. 2020. PMID: 33057196 Free PMC article.
-
The sympathetic nervous system in the 21st century: Neuroimmune interactions in metabolic homeostasis and obesity.Neuron. 2022 Nov 2;110(21):3597-3626. doi: 10.1016/j.neuron.2022.10.017. Neuron. 2022. PMID: 36327900 Free PMC article. Review.
References
-
- Alberts B et al. Essential Cell Biology. (Garland Pub, 2014).
-
- Guo G et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 (2010). - PubMed
-
- Thorsen T, Roberts RW, Arnold FH & Quake SR Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 86, 4163–4166 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

