The Abundant Tegument Protein pUL25 of Human Cytomegalovirus Prevents Proteasomal Degradation of pUL26 and Supports Its Suppression of ISGylation
- PMID: 30282718
- PMCID: PMC6258951
- DOI: 10.1128/JVI.01180-18
The Abundant Tegument Protein pUL25 of Human Cytomegalovirus Prevents Proteasomal Degradation of pUL26 and Supports Its Suppression of ISGylation
Abstract
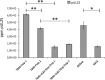
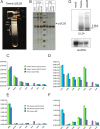
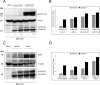
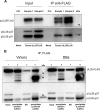
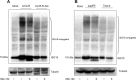
The tegument of human cytomegalovirus (HCMV) virions contains proteins that interfere with both the intrinsic and the innate immunity. One protein with a thus far unknown function is pUL25. The deletion of pUL25 in a viral mutant (Towne-ΔUL25) had no impact on the release of virions and subviral dense bodies or on virion morphogenesis. Proteomic analyses showed few alterations in the overall protein composition of extracellular particles. A surprising result, however, was the almost complete absence of pUL26 in virions and dense bodies of Towne-ΔUL25 and a reduction of the large isoform pUL26-p27 in mutant virus-infected cells. pUL26 had been shown to inhibit protein conjugation with the interferon-stimulated gene 15 protein (ISG15), thereby supporting HCMV replication. To test for a functional relationship between pUL25 and pUL26, we addressed the steady-state levels of pUL26 and found them to be reduced in Towne-ΔUL25-infected cells. Coimmunoprecipitation experiments proved an interaction between pUL25 and pUL26. Surprisingly, the overall protein ISGylation was enhanced in Towne-ΔUL25-infected cells, thus mimicking the phenotype of a pUL26-deleted HCMV mutant. The functional relevance of this was confirmed by showing that the replication of Towne-ΔUL25 was more sensitive to beta interferon. The increase of protein ISGylation was also seen in cells infected with a mutant lacking the tegument protein pp65. Upon retesting, we found that pUL26 degradation was also increased when pp65 was unavailable. Our experiments show that both pUL25 and pp65 regulate pUL26 degradation and the pUL26-dependent reduction of ISGylation and add pUL25 as another HCMV tegument protein that interferes with the intrinsic immunity of the host cell.IMPORTANCE Human cytomegalovirus (HCMV) expresses a number of tegument proteins that interfere with the intrinsic and the innate defense mechanisms of the cell. Initial induction of the interferon-stimulated gene 15 protein (ISG15) and conjugation of proteins with ISG15 (ISGylation) by HCMV infection are subsequently attenuated by the expression of the viral IE1, pUL50, and pUL26 proteins. This study adds pUL25 as another factor that contributes to suppression of ISGylation. The tegument protein interacts with pUL26 and prevents its degradation by the proteasome. By doing this, it supports its restrictive influence on ISGylation. In addition, a lack of pUL25 enhances the levels of free ISG15, indicating that the tegument protein may interfere with the interferon response on levels other than interacting with pUL26. Knowledge obtained in this study widens our understanding of HCMV immune evasion and may also provide a new avenue for the use of pUL25-negative strains for vaccine production.
Keywords: ISG15; ISGylation; cytomegalovirus; dense bodies; pUL25; pUL26; pp65; tegument.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
Transmembrane Protein pUL50 of Human Cytomegalovirus Inhibits ISGylation by Downregulating UBE1L.J Virol. 2018 Jul 17;92(15):e00462-18. doi: 10.1128/JVI.00462-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29743376 Free PMC article.
-
Consecutive Inhibition of ISG15 Expression and ISGylation by Cytomegalovirus Regulators.PLoS Pathog. 2016 Aug 26;12(8):e1005850. doi: 10.1371/journal.ppat.1005850. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27564865 Free PMC article.
-
Dynamic regulatory interaction between cytomegalovirus major tegument protein pp65 and protein kinase pUL97 in intracellular compartments, dense bodies and virions.J Gen Virol. 2017 Nov;98(11):2850-2863. doi: 10.1099/jgv.0.000939. Epub 2017 Oct 12. J Gen Virol. 2017. PMID: 29022869
-
The human cytomegalovirus tegument protein pp65 (pUL83): a key player in innate immune evasion.New Microbiol. 2018 Apr;41(2):87-94. Epub 2018 Jan 31. New Microbiol. 2018. PMID: 29384558 Review.
-
Tegument proteins of human cytomegalovirus.Microbiol Mol Biol Rev. 2008 Jun;72(2):249-65, table of contents. doi: 10.1128/MMBR.00040-07. Microbiol Mol Biol Rev. 2008. PMID: 18535146 Free PMC article. Review.
Cited by
-
Subviral Dense Bodies of Human Cytomegalovirus Induce an Antiviral Type I Interferon Response.Cells. 2022 Dec 13;11(24):4028. doi: 10.3390/cells11244028. Cells. 2022. PMID: 36552792 Free PMC article.
-
HCMV-Mediated Interference of Bortezomib-Induced Apoptosis in Colon Carcinoma Cell Line Caco-2.Viruses. 2021 Jan 9;13(1):83. doi: 10.3390/v13010083. Viruses. 2021. PMID: 33435377 Free PMC article.
-
Recombinant Human Cytomegalovirus Expressing an Analog-Sensitive Kinase pUL97 as Novel Tool for Functional Analyses.Viruses. 2022 Oct 17;14(10):2285. doi: 10.3390/v14102285. Viruses. 2022. PMID: 36298840 Free PMC article.
-
An Attenuated Strain of Human Cytomegalovirus for the Establishment of a Subviral Particle Vaccine.Vaccines (Basel). 2022 Aug 16;10(8):1326. doi: 10.3390/vaccines10081326. Vaccines (Basel). 2022. PMID: 36016214 Free PMC article.
-
Constitutive immune mechanisms: mediators of host defence and immune regulation.Nat Rev Immunol. 2021 Mar;21(3):137-150. doi: 10.1038/s41577-020-0391-5. Epub 2020 Aug 11. Nat Rev Immunol. 2021. PMID: 32782357 Free PMC article. Review.
References
-
- Griffiths PD, Reeves M. 2017. Cytomegalovirus, p 481–510. In Richman D, Whitley RJ, Hayden FG (ed), Clinical virology, 4th ed ASM Press, Washington, DC.
-
- Dai X, Yu X, Gong H, Jiang X, Abenes G, Liu H, Shivakoti S, Britt WJ, Zhu H, Liu F, Zhou ZH. 2013. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog 9:e1003525. doi:10.1371/journal.ppat.1003525. - DOI - PMC - PubMed
-
- Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

