Erythro-myeloid progenitors contribute endothelial cells to blood vessels
- PMID: 30258231
- PMCID: PMC6289247
- DOI: 10.1038/s41586-018-0552-x
Erythro-myeloid progenitors contribute endothelial cells to blood vessels
Abstract
The earliest blood vessels in mammalian embryos are formed when endothelial cells differentiate from angioblasts and coalesce into tubular networks. Thereafter, the endothelium is thought to expand solely by proliferation of pre-existing endothelial cells. Here we show that a complementary source of endothelial cells is recruited into pre-existing vasculature after differentiation from the earliest precursors of erythrocytes, megakaryocytes and macrophages, the erythro-myeloid progenitors (EMPs) that are born in the yolk sac. A first wave of EMPs contributes endothelial cells to the yolk sac endothelium, and a second wave of EMPs colonizes the embryo and contributes endothelial cells to intraembryonic endothelium in multiple organs, where they persist into adulthood. By demonstrating that EMPs constitute a hitherto unrecognized source of endothelial cells, we reveal that embryonic blood vascular endothelium expands in a dual mechanism that involves both the proliferation of pre-existing endothelial cells and the incorporation of endothelial cells derived from haematopoietic precursors.
Conflict of interest statement
The authors declare no competing interests.
Figures


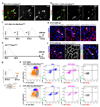


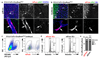
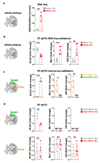
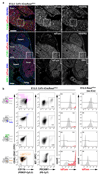






Comment in
-
A dual origin for blood vessels.Nature. 2018 Oct;562(7726):195-197. doi: 10.1038/d41586-018-06199-2. Nature. 2018. PMID: 30291309 Free PMC article.
Similar articles
-
Erythro-myeloid progenitors can differentiate from endothelial cells and modulate embryonic vascular remodeling.Sci Rep. 2017 Mar 8;7:43817. doi: 10.1038/srep43817. Sci Rep. 2017. PMID: 28272478 Free PMC article.
-
No Evidence for Erythro-Myeloid Progenitor-Derived Vascular Endothelial Cells in Multiple Organs.Circ Res. 2020 Oct 23;127(10):1221-1232. doi: 10.1161/CIRCRESAHA.120.317442. Epub 2020 Aug 14. Circ Res. 2020. PMID: 32791884
-
Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors.Nature. 2015 Feb 26;518(7540):547-51. doi: 10.1038/nature13989. Epub 2014 Dec 3. Nature. 2015. PMID: 25470051 Free PMC article.
-
First blood: the endothelial origins of hematopoietic progenitors.Angiogenesis. 2021 May;24(2):199-211. doi: 10.1007/s10456-021-09783-9. Epub 2021 Mar 30. Angiogenesis. 2021. PMID: 33783643 Free PMC article. Review.
-
Fetal monocytes and the origins of tissue-resident macrophages.Cell Immunol. 2018 Aug;330:5-15. doi: 10.1016/j.cellimm.2018.01.001. Epub 2018 Jan 12. Cell Immunol. 2018. PMID: 29475558 Review.
Cited by
-
Angiodiversity and organotypic functions of sinusoidal endothelial cells.Angiogenesis. 2021 May;24(2):289-310. doi: 10.1007/s10456-021-09780-y. Epub 2021 Mar 21. Angiogenesis. 2021. PMID: 33745018 Free PMC article. Review.
-
The Formation of Coronary Vessels in Cardiac Development and Disease.Cold Spring Harb Perspect Biol. 2020 May 1;12(5):a037168. doi: 10.1101/cshperspect.a037168. Cold Spring Harb Perspect Biol. 2020. PMID: 31636078 Free PMC article. Review.
-
An update on clonality: what smooth muscle cell type makes up the atherosclerotic plaque?F1000Res. 2018 Dec 21;7:F1000 Faculty Rev-1969. doi: 10.12688/f1000research.15994.1. eCollection 2018. F1000Res. 2018. PMID: 30613386 Free PMC article. Review.
-
Heat shock protein 60 regulates yolk sac erythropoiesis in mice.Cell Death Dis. 2019 Oct 10;10(10):766. doi: 10.1038/s41419-019-2014-2. Cell Death Dis. 2019. PMID: 31601784 Free PMC article.
-
In vivo generation of haematopoietic stem/progenitor cells from bone marrow-derived haemogenic endothelium.Nat Cell Biol. 2019 Nov;21(11):1334-1345. doi: 10.1038/s41556-019-0410-6. Epub 2019 Nov 4. Nat Cell Biol. 2019. PMID: 31685991
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

