Persistent repair intermediates induce senescence
- PMID: 30254262
- PMCID: PMC6156224
- DOI: 10.1038/s41467-018-06308-9
Persistent repair intermediates induce senescence
Abstract
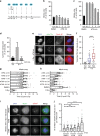
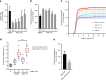
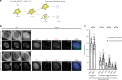
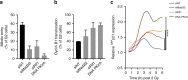
Double-stranded DNA breaks activate a DNA damage checkpoint in G2 phase to trigger a cell cycle arrest, which can be reversed to allow for recovery. However, damaged G2 cells can also permanently exit the cell cycle, going into senescence or apoptosis, raising the question how an individual cell decides whether to recover or withdraw from the cell cycle. Here we find that the decision to withdraw from the cell cycle in G2 is critically dependent on the progression of DNA repair. We show that delayed processing of double strand breaks through HR-mediated repair results in high levels of resected DNA and enhanced ATR-dependent signalling, allowing p21 to rise to levels at which it drives cell cycle exit. These data imply that cells have the capacity to discriminate breaks that can be repaired from breaks that are difficult to repair at a time when repair is still ongoing.
Conflict of interest statement
The authors declare no competing interests
Figures

Similar articles
-
Chk1 is dispensable for G2 arrest in response to sustained DNA damage when the ATM/p53/p21 pathway is functional.Oncogene. 2011 Oct 13;30(41):4261-74. doi: 10.1038/onc.2011.135. Epub 2011 May 2. Oncogene. 2011. PMID: 21532626
-
Residual Cdk1/2 activity after DNA damage promotes senescence.Aging Cell. 2017 Jun;16(3):575-584. doi: 10.1111/acel.12588. Epub 2017 Mar 26. Aging Cell. 2017. PMID: 28345297 Free PMC article.
-
Nuclear translocation of Cyclin B1 marks the restriction point for terminal cell cycle exit in G2 phase.Cell Cycle. 2014;13(17):2733-43. doi: 10.4161/15384101.2015.945831. Cell Cycle. 2014. PMID: 25486360 Free PMC article.
-
DNA damage responses in skin biology--implications in tumor prevention and aging acceleration.J Dermatol Sci. 2009 Nov;56(2):76-81. doi: 10.1016/j.jdermsci.2009.09.001. Epub 2009 Sep 24. J Dermatol Sci. 2009. PMID: 19781914 Review.
-
[Cell cycle regulation after exposure to ionizing radiation].Bull Cancer. 1999 Apr;86(4):345-57. Bull Cancer. 1999. PMID: 10341340 Review. French.
Cited by
-
The role of Sp1 in the detection and elimination of cells with persistent DNA strand breaks.NAR Cancer. 2020 Mar 14;2(2):zcaa004. doi: 10.1093/narcan/zcaa004. eCollection 2020 Jun. NAR Cancer. 2020. PMID: 34316684 Free PMC article.
-
Sublethal engagement of apoptotic pathways in residual cancer.Trends Cell Biol. 2024 Mar;34(3):225-238. doi: 10.1016/j.tcb.2023.07.005. Epub 2023 Aug 10. Trends Cell Biol. 2024. PMID: 37573235 Review.
-
Spatiotemporal Mislocalization of Nuclear Membrane-Associated Proteins in γ-Irradiation-Induced Senescent Cells.Cells. 2020 Apr 17;9(4):999. doi: 10.3390/cells9040999. Cells. 2020. PMID: 32316379 Free PMC article.
-
DNA Topoisomerase 3α Is Involved in Homologous Recombination Repair and Replication Stress Response in Trypanosoma cruzi.Front Cell Dev Biol. 2021 May 13;9:633195w. doi: 10.3389/fcell.2021.633195. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34055812 Free PMC article.
-
Heterogenous Differences in Cellular Senescent Phenotypes in Pre-Eclampsia and IUGR following Quantitative Assessment of Multiple Biomarkers of Senescence.Int J Mol Sci. 2023 Feb 4;24(4):3101. doi: 10.3390/ijms24043101. Int J Mol Sci. 2023. PMID: 36834513 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

