N-terminal acetylation and methylation differentially affect the function of MYL9
- PMID: 30242065
- PMCID: PMC6442934
- DOI: 10.1042/BCJ20180638
N-terminal acetylation and methylation differentially affect the function of MYL9
Abstract
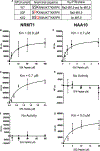
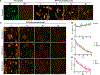
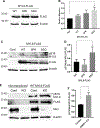
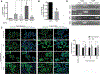
Deciphering the histone code has illustrated that acetylation or methylation on the same residue can have analogous or opposing roles. However, little is known about the interplay between these post-translational modifications (PTMs) on the same nonhistone residues. We have recently discovered that N-terminal acetyltransferases (NATs) and N-terminal methyltransferases (NRMTs) can have overlapping substrates and identified myosin regulatory light chain 9 (MYL9) as the first confirmed protein to occur in either α-amino-methylated (Nα-methyl) or α-amino-acetylated (Nα-acetyl) states in vivo Here we aim to determine if these PTMs function similarly or create different MYL9 proteoforms with distinct roles. We use enzymatic assays to directly verify MYL9 is a substrate of both NRMT1 and NatA and generate mutants of MYL9 that are exclusive for Nα-acetylation or Nα-methylation. We then employ eukaryotic cell models to probe the regulatory functions of these Nα-PTMs on MYL9. Our results show that, contrary to prevailing dogma, neither of these modifications regulate the stability of MYL9. Rather, exclusive Nα-acetylation promotes cytoplasmic roles of MYL9, while exclusive Nα-methylation promotes the nuclear role of MYL9 as a transcription factor. The increased cytoplasmic activity of Nα-acetylated MYL9 corresponds with increased phosphorylation at serine 19, a key MYL9 activating PTM. Increased nuclear activity of Nα-methylated MYL9 corresponds with increased DNA binding. Nα-methylation also results in a decrease of interactions between the N-terminus of MYL9 and a host of cytoskeletal proteins. These results confirm that Nα-acetylation and Nα-methylation differentially affect MYL9 function by creating distinct proteoforms with different internal PTM patterns and binding properties.
Keywords: acetylation/deacetylation; methylation; myosins.
© 2018 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Figures


Similar articles
-
A novel human NatA Nalpha-terminal acetyltransferase complex: hNaa16p-hNaa10p (hNat2-hArd1).BMC Biochem. 2009 May 29;10:15. doi: 10.1186/1471-2091-10-15. BMC Biochem. 2009. PMID: 19480662 Free PMC article.
-
Three's a crowd - why did three N-terminal methyltransferases evolve for one job?J Cell Sci. 2023 Jan 15;136(2):jcs260424. doi: 10.1242/jcs.260424. Epub 2023 Jan 17. J Cell Sci. 2023. PMID: 36647772 Free PMC article.
-
Characterizations of Protein Arginine Deiminase 1 as a Substrate of NTMT1: Implications of Nα-Methylation in Protein Stability and Interaction.J Proteome Res. 2024 Oct 4;23(10):4589-4600. doi: 10.1021/acs.jproteome.4c00484. Epub 2024 Sep 17. J Proteome Res. 2024. PMID: 39287128
-
Unlocking the mysteries of alpha-N-terminal methylation and its diverse regulatory functions.J Biol Chem. 2023 Jul;299(7):104843. doi: 10.1016/j.jbc.2023.104843. Epub 2023 May 19. J Biol Chem. 2023. PMID: 37209820 Free PMC article. Review.
-
N-terminal acetylation and other functions of Nα-acetyltransferases.Biol Chem. 2012 Apr;393(4):291-8. doi: 10.1515/hsz-2011-0228. Biol Chem. 2012. PMID: 22718636 Review.
Cited by
-
Intrafamily heterooligomerization as an emerging mechanism of methyltransferase regulation.Epigenetics Chromatin. 2024 Mar 1;17(1):5. doi: 10.1186/s13072-024-00530-0. Epigenetics Chromatin. 2024. PMID: 38429855 Free PMC article. Review.
-
Design methods for antimicrobial peptides with improved performance.Zool Res. 2023 Nov 18;44(6):1095-1114. doi: 10.24272/j.issn.2095-8137.2023.246. Zool Res. 2023. PMID: 37914524 Free PMC article. Review.
-
Design and characterization of PROTAC degraders specific to protein N-terminal methyltransferase 1.Eur J Med Chem. 2022 Dec 15;244:114830. doi: 10.1016/j.ejmech.2022.114830. Epub 2022 Oct 5. Eur J Med Chem. 2022. PMID: 36228414 Free PMC article.
-
A Strong Cation Exchange Chromatography Protocol for Examining N-Terminal Proteoforms.Methods Mol Biol. 2022;2477:293-309. doi: 10.1007/978-1-0716-2257-5_17. Methods Mol Biol. 2022. PMID: 35524124
-
Novel regulation of the transcription factor ZHX2 by N-terminal methylation.Transcription. 2022 Feb-Jun;13(1-3):1-15. doi: 10.1080/21541264.2022.2079184. Epub 2022 May 25. Transcription. 2022. PMID: 35613330 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

