A whole-blood RNA transcript-based gene signature is associated with the development of CTLA-4 blockade-related diarrhea in patients with advanced melanoma treated with the checkpoint inhibitor tremelimumab
- PMID: 30227886
- PMCID: PMC6145108
- DOI: 10.1186/s40425-018-0408-9
A whole-blood RNA transcript-based gene signature is associated with the development of CTLA-4 blockade-related diarrhea in patients with advanced melanoma treated with the checkpoint inhibitor tremelimumab
Abstract
Background: Anti-CTLA-4 immune checkpoint blockade is associated with immune-related adverse events (irAEs). Grade 3-4 diarrhea/colitis is the most frequent irAE requiring treatment discontinuation. Predicting high-risk diarrhea/colitis patients may facilitate early intervention, limit irAE severity, and extend treatment duration. No biomarkers currently predict for anti-CTLA-4 immunotherapy related severe diarrhea.
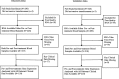
Methods: Whole-blood was collected pre-treatment and 30 days post-treatment initiation from patients with stage III or IV unresectable melanoma who received 15 mg/kg tremelimumab at 90 day intervals in two clinical trials. The discovery dataset was a phase II study that enrolled 150 patients between December 2005 and November 2006. The validation dataset was a phase III study that enrolled 210 patients between March 2006 and July 2007. RT-PCR was performed for 169 genes associated with inflammation, immunity, CTLA-4 pathway and melanoma. Gene expression was correlated with grade 0-1 versus grade 2-4 diarrhea/colitis development.
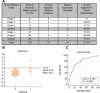
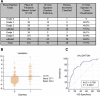
Results: Pre-treatment blood obtained from the discovery dataset (N = 150) revealed no gene predictive of diarrhea/colitis development (p < 0.05). A 16-gene signature (CARD12, CCL3, CCR3, CXCL1, F5, FAM210B, GADD45A, IL18bp, IL2RA, IL5, IL8, MMP9, PTGS2, SOCS3, TLR9 and UBE2C) was identified from 30 days post-tremelimumab initiation blood that discriminated patients developing grade 0-1 from grade 2-4 diarrhea/colitis. The 16-gene signature demonstrated an AUC of 0.814 (95% CI 0.743 to 0.873, p < 0.0001), sensitivity 42.9%, specificity 99.2%, positive predictive value (PPV) 90.0%, and negative predictive value (NPV) 91.4%. In the validation dataset (N = 210), the 16-gene signature discriminated patients developing grade 0-1 from grade 2-4 diarrhea/colitis with an AUC 0.785 (95% CI 0.723 to 0.838, p < 0.0001), sensitivity 57.1%, specificity 84.4%, PPV 57.1% and NPV 84.4%.
Conclusion: This study identifies a whole-blood mRNA signature predictive of a clinically relevant irAE in patients treated with immune checkpoint blockade. We hypothesize that immune system modulation induced by immune checkpoint blockade results in peripheral blood gene expression changes that are detectable prior to clinical onset of severe diarrhea. Assessment of peripheral blood gene expression changes in patients receiving anti-PD-1/PD-L1 immunotherapy, or combination anti-CTLA4 and anti-PD-1/PD-L1 immunotherapy, is warranted to provide early on-treatment mechanistic insights and identify clinically relevant predictive biomarkers.
Trial registration: Clinicaltrials.gov , NCT00257205 , registered 22 November 2005.
Keywords: Biomarker; CTLA-4; Diarrhea; Immunotherapy; Melanoma; Predictive; Tremelimumab; irAE.
Conflict of interest statement
Ethics approval and consent to participate
Separate ethical approval is not needed for the current submission. This manuscript presents data analyzing RNA obtained from blood of patients enrolled in two different Pfizer sponsored clinical trials. RNA analysis of the Pfizer study data was contemplated in the original protocol consents and therefore this study does not need ethical approval as the subjects had consented to participate in the two studies and agreed to have their blood drawn and gene expression analyzed. Furthermore the investigators remain totally blinded as to the patient’s identifiers with HIPPA protected data.
Consent for publication
not applicable.
Competing interests
PF: Advisory Board Member for Seattle Genetics, Regeneron, Aspyrian Therapeutics, Array Biopharma, EMD Serono and Castle Biosciences. Stock equity in MACK, AGN, INCY, ADVM, and CLVS.
KW M.D.: no conflicts.
KW: President and CEO of CPS Companion Diagnostics. Equity interest in CPS Companion Diagnostics which holds patent related to content of the manuscript.
AMC: Chairman of CPS Companion Diagnostics. Equity interest in CPS Companion Diagnostics which holds patent related to content of the manuscript.
NB: Consultant for Neon. Advisory Board Member for CPS Companion Diagnostics and Curevac. Equity interest in CPS Companion Diagnostics which holds patent related to content of the manuscript.
WKO: Chief Scientific Officer and Chairman of the Science Advisory Board CPS Companion Diagnostics. Equity interest in CPS Companion Diagnostics which holds patent related to content of the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Whole-blood RNA transcript-based models can predict clinical response in two large independent clinical studies of patients with advanced melanoma treated with the checkpoint inhibitor, tremelimumab.J Immunother Cancer. 2017 Aug 15;5(1):67. doi: 10.1186/s40425-017-0272-z. J Immunother Cancer. 2017. PMID: 28807052 Free PMC article. Clinical Trial.
-
The Risk of Diarrhea and Colitis in Patients With Advanced Melanoma Undergoing Immune Checkpoint Inhibitor Therapy: A Systematic Review and Meta-Analysis.J Immunother. 2018 Apr;41(3):101-108. doi: 10.1097/CJI.0000000000000213. J Immunother. 2018. PMID: 29401166
-
Blood mRNA expression profiling predicts survival in patients treated with tremelimumab.Clin Cancer Res. 2014 Jun 15;20(12):3310-8. doi: 10.1158/1078-0432.CCR-13-2906. Epub 2014 Apr 10. Clin Cancer Res. 2014. PMID: 24721645 Clinical Trial.
-
The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma.Clin Ther. 2015 Apr 1;37(4):764-82. doi: 10.1016/j.clinthera.2015.02.018. Epub 2015 Mar 29. Clin Ther. 2015. PMID: 25823918 Free PMC article. Review.
-
Therapeutic use of anti-CTLA-4 antibodies.Int Immunol. 2015 Jan;27(1):3-10. doi: 10.1093/intimm/dxu076. Epub 2014 Jul 18. Int Immunol. 2015. PMID: 25038057 Review.
Cited by
-
Pneumonitis resulting from radiation and immune checkpoint blockade illustrates characteristic clinical, radiologic and circulating biomarker features.J Immunother Cancer. 2019 Apr 24;7(1):112. doi: 10.1186/s40425-019-0583-3. J Immunother Cancer. 2019. PMID: 31014385 Free PMC article.
-
Immune-related toxicities of checkpoint inhibitors: mechanisms and mitigation strategies.Nat Rev Drug Discov. 2022 Jul;21(7):495-508. doi: 10.1038/s41573-021-00259-5. Epub 2021 Jul 27. Nat Rev Drug Discov. 2022. PMID: 34316029 Review.
-
Predictive Biomarkers of Immune Checkpoint Inhibitors-Related Toxicities.Front Immunol. 2020 Oct 6;11:2023. doi: 10.3389/fimmu.2020.02023. eCollection 2020. Front Immunol. 2020. PMID: 33123120 Free PMC article. Review.
-
Whole Blood Transcriptome Profiling Identifies DNA Replication and Cell Cycle Regulation as Early Marker of Response to Anti-PD-1 in Patients with Urothelial Cancer.Cancers (Basel). 2021 Sep 17;13(18):4660. doi: 10.3390/cancers13184660. Cancers (Basel). 2021. PMID: 34572887 Free PMC article.
-
Factors associated with immune‑related severe adverse events (Review).Mol Clin Oncol. 2024 Oct 31;22(1):3. doi: 10.3892/mco.2024.2798. eCollection 2025 Jan. Mol Clin Oncol. 2024. PMID: 39563998 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
