The Nucleolar Protein LYAR Facilitates Ribonucleoprotein Assembly of Influenza A Virus
- PMID: 30209172
- PMCID: PMC6232469
- DOI: 10.1128/JVI.01042-18
The Nucleolar Protein LYAR Facilitates Ribonucleoprotein Assembly of Influenza A Virus
Abstract
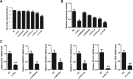
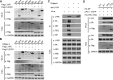
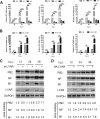
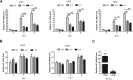
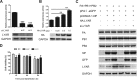
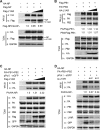
Influenza A viral ribonucleoprotein (vRNP) is responsible for transcription and replication of the viral genome in infected cells and depends on host factors for its functions. Identification of the host factors interacting with vRNP not only improves understanding of virus-host interactions but also provides insights into novel mechanisms of viral pathogenicity and the development of new antiviral strategies. Here, we have identified 80 host factors that copurified with vRNP using affinity purification followed by mass spectrometry. LYAR, a cell growth-regulating nucleolar protein, has been shown to be important for influenza A virus replication. During influenza A virus infection, LYAR expression is increased and partly translocates from the nucleolus to the nucleoplasm and cytoplasm. Furthermore, LYAR interacts with RNP subunits, resulting in enhancing viral RNP assembly, thereby facilitating viral RNA synthesis. Taken together, our studies identify a novel vRNP binding host partner important for influenza A virus replication and further reveal the mechanism of LYAR regulating influenza A viral RNA synthesis by facilitating viral RNP assembly.IMPORTANCE Influenza A virus (IAV) must utilize the host cell machinery to replicate, but many of the mechanisms of IAV-host interaction remain poorly understood. Improved understanding of interactions between host factors and vRNP not only increases our basic knowledge of the molecular mechanisms of virus replication and pathogenicity but also provides insights into possible novel antiviral targets that are necessary due to the widespread emergence of drug-resistant IAV strains. Here, we have identified LYAR, a cell growth-regulating nucleolar protein, which interacts with viral RNP components and is important for efficient replication of IAVs and whose role in the IAV life cycle has never been reported. In addition, we further reveal the role of LYAR in viral RNA synthesis. Our results extend and improve current knowledge on the mechanisms of IAV transcription and replication.
Keywords: LYAR; host factor; influenza A virus; vRNP; viral RNP assembly.
Copyright © 2018 Yang et al.
Figures

Similar articles
-
Eukaryotic Translation Elongation Factor 1 Delta Inhibits the Nuclear Import of the Nucleoprotein and PA-PB1 Heterodimer of Influenza A Virus.J Virol. 2020 Dec 22;95(2):e01391-20. doi: 10.1128/JVI.01391-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087462 Free PMC article.
-
Y-Box-Binding Protein 3 (YBX3) Restricts Influenza A Virus by Interacting with Viral Ribonucleoprotein Complex and Imparing its Function.J Gen Virol. 2020 Apr;101(4):385-398. doi: 10.1099/jgv.0.001390. J Gen Virol. 2020. PMID: 32553055
-
Autophagy Promotes Replication of Influenza A Virus In Vitro.J Virol. 2019 Feb 5;93(4):e01984-18. doi: 10.1128/JVI.01984-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30541828 Free PMC article.
-
Structure and Function of the Influenza Virus Transcription and Replication Machinery.Cold Spring Harb Perspect Med. 2020 Sep 1;10(9):a038398. doi: 10.1101/cshperspect.a038398. Cold Spring Harb Perspect Med. 2020. PMID: 31871230 Free PMC article. Review.
-
Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza A virus infected cell.RNA Biol. 2013 Aug;10(8):1274-82. doi: 10.4161/rna.25356. Epub 2013 Jun 17. RNA Biol. 2013. PMID: 23807439 Free PMC article. Review.
Cited by
-
Chasing Intracellular Zika Virus Using Proteomics.Viruses. 2019 Sep 19;11(9):878. doi: 10.3390/v11090878. Viruses. 2019. PMID: 31546825 Free PMC article. Review.
-
Nonmuscle myosin IIA promotes the internalization of influenza A virus and regulates viral polymerase activity through interacting with nucleoprotein in human pulmonary cells.Virol Sin. 2023 Feb;38(1):128-141. doi: 10.1016/j.virs.2022.12.002. Epub 2022 Dec 10. Virol Sin. 2023. PMID: 36509386 Free PMC article.
-
Modification of Nuclear Compartments and the 3D Genome in the Course of a Viral Infection.Acta Naturae. 2020 Oct-Dec;12(4):34-46. doi: 10.32607/actanaturae.11041. Acta Naturae. 2020. PMID: 33456976 Free PMC article.
-
Transcriptomic response to ISAV infection in the gills, head kidney and spleen of resistant and susceptible Atlantic salmon.BMC Genomics. 2022 Nov 28;23(1):775. doi: 10.1186/s12864-022-09007-4. BMC Genomics. 2022. PMID: 36443659 Free PMC article.
-
Nucleoporin 85 interacts with influenza A virus PB1 and PB2 to promote its replication by facilitating nuclear import of ribonucleoprotein.Front Microbiol. 2022 Aug 16;13:895779. doi: 10.3389/fmicb.2022.895779. eCollection 2022. Front Microbiol. 2022. PMID: 36051755 Free PMC article.
References
-
- . 2013. Infectious Disease/CDC Update: Update on emerging infections: news from the Centers for Disease Control and Prevention. Evaluation of 11 commercially available rapid influenza diagnostic tests-United States, 2011-2012. Ann Emerg Med 61:573–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

