Genomic and epidemiological monitoring of yellow fever virus transmission potential
- PMID: 30139911
- PMCID: PMC6874500
- DOI: 10.1126/science.aat7115
Genomic and epidemiological monitoring of yellow fever virus transmission potential
Abstract
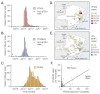
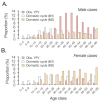
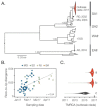
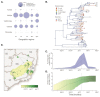
The yellow fever virus (YFV) epidemic in Brazil is the largest in decades. The recent discovery of YFV in Brazilian Aedes species mosquitos highlights a need to monitor the risk of reestablishment of urban YFV transmission in the Americas. We use a suite of epidemiological, spatial, and genomic approaches to characterize YFV transmission. We show that the age and sex distribution of human cases is characteristic of sylvatic transmission. Analysis of YFV cases combined with genomes generated locally reveals an early phase of sylvatic YFV transmission and spatial expansion toward previously YFV-free areas, followed by a rise in viral spillover to humans in late 2016. Our results establish a framework for monitoring YFV transmission in real time that will contribute to a global strategy to eliminate future YFV epidemics.
Copyright © 2018, American Association for the Advancement of Science.
Conflict of interest statement
N.J.L. and L.C.J.A. received free-of-charge reagents in support of the project from Oxford Nanopore Technologies.
Figures
Comment in
-
The reemergence of yellow fever.Science. 2018 Aug 31;361(6405):847-848. doi: 10.1126/science.aau8225. Epub 2018 Aug 23. Science. 2018. PMID: 30139914 No abstract available.
-
Monitoring yellow fever.Nat Med. 2018 Dec;24(12):1781. doi: 10.1038/s41591-018-0280-7. Nat Med. 2018. PMID: 30523315 No abstract available.
Similar articles
-
Yellow Fever Virus Reemergence and Spread in Southeast Brazil, 2016-2019.J Virol. 2019 Dec 12;94(1):e01623-19. doi: 10.1128/JVI.01623-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597773 Free PMC article.
-
Persistence of Yellow fever virus outside the Amazon Basin, causing epidemics in Southeast Brazil, from 2016 to 2018.PLoS Negl Trop Dis. 2018 Jun 4;12(6):e0006538. doi: 10.1371/journal.pntd.0006538. eCollection 2018 Jun. PLoS Negl Trop Dis. 2018. PMID: 29864115 Free PMC article.
-
Neighbor danger: Yellow fever virus epizootics in urban and urban-rural transition areas of Minas Gerais state, during 2017-2018 yellow fever outbreaks in Brazil.PLoS Negl Trop Dis. 2020 Oct 5;14(10):e0008658. doi: 10.1371/journal.pntd.0008658. eCollection 2020 Oct. PLoS Negl Trop Dis. 2020. PMID: 33017419 Free PMC article.
-
Recent sylvatic yellow fever virus transmission in Brazil: the news from an old disease.Virol J. 2020 Jan 23;17(1):9. doi: 10.1186/s12985-019-1277-7. Virol J. 2020. PMID: 31973727 Free PMC article. Review.
-
Yellow fever outbreak in Brazil: the puzzle of rapid viral spread and challenges for immunisation.Mem Inst Oswaldo Cruz. 2018 Sep 3;113(10):e180278. doi: 10.1590/0074-02760180278. Mem Inst Oswaldo Cruz. 2018. PMID: 30427974 Free PMC article. Review.
Cited by
-
Field and classroom initiatives for portable sequence-based monitoring of dengue virus in Brazil.Nat Commun. 2021 Apr 16;12(1):2296. doi: 10.1038/s41467-021-22607-0. Nat Commun. 2021. PMID: 33863880 Free PMC article.
-
Investigation of the validity of two Bayesian ancestral state reconstruction models for estimating Salmonella transmission during outbreaks.PLoS One. 2019 Jul 22;14(7):e0214169. doi: 10.1371/journal.pone.0214169. eCollection 2019. PLoS One. 2019. PMID: 31329588 Free PMC article.
-
An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar.Genome Biol. 2019 Jan 8;20(1):8. doi: 10.1186/s13059-018-1618-7. Genome Biol. 2019. PMID: 30621750 Free PMC article.
-
Circulation of chikungunya virus East/Central/South African lineage in Rio de Janeiro, Brazil.PLoS One. 2019 Jun 11;14(6):e0217871. doi: 10.1371/journal.pone.0217871. eCollection 2019. PLoS One. 2019. PMID: 31185030 Free PMC article. Clinical Trial.
-
West Nile virus transmission potential in Portugal.Commun Biol. 2022 Jan 10;5(1):6. doi: 10.1038/s42003-021-02969-3. Commun Biol. 2022. PMID: 35013546 Free PMC article.
References
-
- Paules CI, Fauci AS. Yellow Fever - Once Again on the Radar Screen in the Americas. The New England Journal of Medicine. 2017:1397–1399. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

