Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes
- PMID: 30139387
- PMCID: PMC6107963
- DOI: 10.1186/s40246-018-0173-3
Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes
Abstract
Background: Studying epigenetics is expected to provide precious information on how environmental factors contribute to type 2 diabetes mellitus (T2DM) at the genomic level. With the progress of the whole-genome resequencing efforts, it is now known that 75-90% of the human genome was transcribed to generate a series of long non-coding RNAs (lncRNAs). While lncRNAs are gaining widespread attention as potential and robust biomarkers in the genesis as well as progression of several disease states, their clinical relevance and regulatory mechanisms are yet to be explored in the field of metabolic disorders including diabetes. Despite the fact that Asian Indians are highly insulin resistant and more prone to develop T2DM and associated vascular complications, there is virtually lack of data on the role of lncRNAs in the clinical diabetes setting. Therefore, we sought to evaluate a panel of lncRNAs and senescence-inflammation signatures in peripheral blood mononuclear cells (PBMCs) from patients with type 2 diabetes (T2DM; n = 30) compared to individuals with normal glucose tolerance (NGT; n = 32).
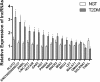
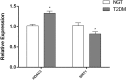
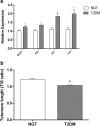
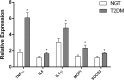
Results: Compared to control subjects, expression levels of lncRNAs in PBMCs from type 2 diabetes patients showed significantly (p < 0.05) increased levels of HOTAIR, MEG3, LET, MALAT1, MIAT, CDKN2BAS1/ANRIL, XIST, PANDA, GAS5, Linc-p21, ENST00000550337.1, PLUTO, and NBR2. In contrast, lncRNA expression patterns of THRIL and SALRNA1 were significantly (p < 0.05) decreased in patients with T2DM compared to control subjects. At the transcriptional level, senescence markers (p53, p21, p16, and β-galactosidase), proinflammatory markers (TNF-α, IL6, MCP1, and IL1-β), and epigenetic signature of histone deacetylase-3 (HDAC3) were significantly (p < 0.05) elevated in patients with type 2 diabetes compared to control subjects. Interestingly, mRNA expression of Sirt1 and telomere length were significantly (p < 0.05) decreased in patients with type 2 diabetes compared to control subjects. Majority of the altered lncRNAs were positively correlated with poor glycemic control, insulin resistance, transcriptional markers of senescence, inflammation, and HDAC3 and negatively correlated with telomere length. Logistic regression analysis revealed a significant association of altered lncRNA signatures with T2DM, but this association was lost after adjusting for insulin resistance (HOMA-IR) and senescence markers.
Conclusion: Our study provides a clinically relevant evidence for the association of altered lncRNAs with poor glycemic control, insulin resistance, accelerated cellular senescence, and inflammation.
Keywords: HDAC3; Inflammation; Insulin resistance; SASP; Type 2 diabetes; lncRNA.
Conflict of interest statement
Ethics approval and consent to participate
The study was approved by the institutional ethics committee of the Madras Diabetes Research Foundation and conducted according to the principles of Declaration of Helsinki. Written informed consent was obtained from all the study participants prior to the start of the study.
Consent for publication
Institutional consent form is inclusive of data protection and consent for research publication.
All the authors approved the manuscript and consented for publication.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Novel insights of elevated systemic levels of bisphenol-A (BPA) linked to poor glycemic control, accelerated cellular senescence and insulin resistance in patients with type 2 diabetes.Mol Cell Biochem. 2019 Aug;458(1-2):171-183. doi: 10.1007/s11010-019-03540-9. Epub 2019 Apr 20. Mol Cell Biochem. 2019. PMID: 31004310 Clinical Trial.
-
Augmentation of histone deacetylase 3 (HDAC3) epigenetic signature at the interface of proinflammation and insulin resistance in patients with type 2 diabetes.Clin Epigenetics. 2016 Nov 24;8:125. doi: 10.1186/s13148-016-0293-3. eCollection 2016. Clin Epigenetics. 2016. PMID: 27904654 Free PMC article.
-
Long non-coding RNA LY86-AS1 and HCG27_201 expression in type 2 diabetes mellitus.Mol Biol Rep. 2018 Dec;45(6):2601-2608. doi: 10.1007/s11033-018-4429-8. Epub 2018 Oct 16. Mol Biol Rep. 2018. PMID: 30328000
-
Long non-coding RNAs: From disease code to drug role.Acta Pharm Sin B. 2021 Feb;11(2):340-354. doi: 10.1016/j.apsb.2020.10.001. Epub 2020 Oct 10. Acta Pharm Sin B. 2021. PMID: 33643816 Free PMC article. Review.
-
Long noncoding RNAs: Novel insights into hepatocelluar carcinoma.Cancer Lett. 2014 Mar 1;344(1):20-27. doi: 10.1016/j.canlet.2013.10.021. Epub 2013 Oct 30. Cancer Lett. 2014. PMID: 24183851 Review.
Cited by
-
Long Noncoding RNAs and Their Therapeutic Promise in Diabetic Nephropathy.Nephron. 2021;145(4):404-414. doi: 10.1159/000515422. Epub 2021 Apr 14. Nephron. 2021. PMID: 33853077 Free PMC article. Review.
-
Phytochemicals as Potential Epidrugs in Type 2 Diabetes Mellitus.Front Endocrinol (Lausanne). 2021 Jun 1;12:656978. doi: 10.3389/fendo.2021.656978. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34140928 Free PMC article. Review.
-
Insulin Resistance and Cancer: In Search for a Causal Link.Int J Mol Sci. 2021 Oct 15;22(20):11137. doi: 10.3390/ijms222011137. Int J Mol Sci. 2021. PMID: 34681797 Free PMC article. Review.
-
hsa-miR-607, lncRNA TUG1 and hsa_circ_0071106 can be combined as biomarkers in type 2 diabetes mellitus.Exp Biol Med (Maywood). 2022 Sep;247(18):1609-1618. doi: 10.1177/15353702221110648. Epub 2022 Jul 23. Exp Biol Med (Maywood). 2022. PMID: 35876150 Free PMC article.
-
Long Noncoding RNAs and Mitochondrial Homeostasis in the Development of Diabetic Retinopathy.Front Endocrinol (Lausanne). 2022 Jun 6;13:915031. doi: 10.3389/fendo.2022.915031. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35733767 Free PMC article. Review.
References
-
- International Diabetes Federation . IDF diabetes atlas. 8. Brussels: International Diabetes Federation; 2017. - PubMed
-
- Monickaraj F, Aravind S, Gokulakrishnan K, Sathishkumar C, Prabu P, Prabu D, Mohan V, Balasubramanyam M. Accelerated aging as evidenced by increased telomere shortening and mitochondrial DNA depletion in patients with type 2 diabetes. Mol Cell Biochem. 2012;365:343–350. doi: 10.1007/s11010-012-1276-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

