N-glycanase NGLY1 regulates mitochondrial homeostasis and inflammation through NRF1
- PMID: 30135079
- PMCID: PMC6170171
- DOI: 10.1084/jem.20180783
N-glycanase NGLY1 regulates mitochondrial homeostasis and inflammation through NRF1
Abstract
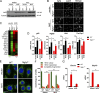
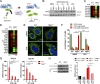
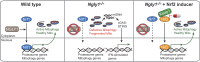
Mutations in the NGLY1 (N-glycanase 1) gene, encoding an evolutionarily conserved deglycosylation enzyme, are associated with a rare congenital disorder leading to global developmental delay and neurological abnormalities. The molecular mechanism of the NGLY1 disease and its function in tissue and immune homeostasis remain unknown. Here, we find that NGLY1-deficient human and mouse cells chronically activate cytosolic nucleic acid-sensing pathways, leading to elevated interferon gene signature. We also find that cellular clearance of damaged mitochondria by mitophagy is impaired in the absence of NGLY1, resulting in severely fragmented mitochondria and activation of cGAS-STING as well as MDA5-MAVS pathways. Furthermore, we show that NGLY1 regulates mitochondrial homeostasis through transcriptional factor NRF1. Remarkably, pharmacological activation of a homologous but nonglycosylated transcriptional factor NRF2 restores mitochondrial homeostasis and suppresses immune gene activation in NGLY1-deficient cells. Together, our findings reveal novel functions of the NGLY1-NRF1 pathway in mitochondrial homeostasis and inflammation and uncover an unexpected therapeutic strategy using pharmacological activators of NRF2 for treating mitochondrial and immune dysregulation.
© 2018 Yang et al.
Figures


Similar articles
-
Drug screens of NGLY1 deficiency in worm and fly models reveal catecholamine, NRF2 and anti-inflammatory-pathway activation as potential clinical approaches.Dis Model Mech. 2019 Nov 4;12(11):dmm040576. doi: 10.1242/dmm.040576. Dis Model Mech. 2019. PMID: 31615832 Free PMC article.
-
N-Glycanase 1 Transcriptionally Regulates Aquaporins Independent of Its Enzymatic Activity.Cell Rep. 2019 Dec 24;29(13):4620-4631.e4. doi: 10.1016/j.celrep.2019.11.097. Cell Rep. 2019. PMID: 31875565
-
Mitochondrial function requires NGLY1.Mitochondrion. 2018 Jan;38:6-16. doi: 10.1016/j.mito.2017.07.008. Epub 2017 Jul 25. Mitochondrion. 2018. PMID: 28750948 Free PMC article.
-
NGLY1: insights from Caenorhabditis elegans.J Biochem. 2022 Feb 21;171(2):145-152. doi: 10.1093/jb/mvab112. J Biochem. 2022. PMID: 34697631 Review.
-
Comprehensive Analysis of the Structure and Function of Peptide:N-Glycanase 1 and Relationship with Congenital Disorder of Deglycosylation.Nutrients. 2022 Apr 19;14(9):1690. doi: 10.3390/nu14091690. Nutrients. 2022. PMID: 35565658 Free PMC article. Review.
Cited by
-
N-glycoproteomics reveals distinct glycosylation alterations in NGLY1-deficient patient-derived dermal fibroblasts.J Inherit Metab Dis. 2023 Jan;46(1):76-91. doi: 10.1002/jimd.12557. Epub 2022 Oct 4. J Inherit Metab Dis. 2023. PMID: 36102038 Free PMC article.
-
DDI2 protease controls embryonic development and inflammation via TCF11/NRF1.iScience. 2024 Sep 5;27(10):110893. doi: 10.1016/j.isci.2024.110893. eCollection 2024 Oct 18. iScience. 2024. PMID: 39328932 Free PMC article.
-
Orthopaedic phenotyping of NGLY1 deficiency using an international, family-led disease registry.Orphanet J Rare Dis. 2019 Jun 19;14(1):148. doi: 10.1186/s13023-019-1131-4. Orphanet J Rare Dis. 2019. PMID: 31217022 Free PMC article.
-
Unexplained death in patients with NGLY1 mutations may be explained by adrenal insufficiency.Physiol Rep. 2019 Feb;7(3):e13979. doi: 10.14814/phy2.13979. Physiol Rep. 2019. PMID: 30740912 Free PMC article.
-
DNA-sensing pathways in health, autoinflammatory and autoimmune diseases.Nat Immunol. 2024 Nov;25(11):2001-2014. doi: 10.1038/s41590-024-01966-y. Epub 2024 Oct 4. Nat Immunol. 2024. PMID: 39367124 Review.
References
-
- Aguirre S., Luthra P., Sanchez-Aparicio M.T., Maestre A.M., Patel J., Lamothe F., Fredericks A.C., Tripathi S., Zhu T., Pintado-Silva J., et al. . 2017. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2:17037 10.1038/nmicrobiol.2017.37 - DOI - PMC - PubMed
-
- Bialas A.R., Presumey J., Das A., van der Poel C.E., Lapchak P.H., Mesin L., Victora G., Tsokos G.C., Mawrin C., Herbst R., and Carroll M.C.. 2017. Microglia-dependent synapse loss in type I interferon-mediated lupus. Nature. 546:539–543. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

