Metronidazole Causes Skeletal Muscle Atrophy and Modulates Muscle Chronometabolism
- PMID: 30115857
- PMCID: PMC6121908
- DOI: 10.3390/ijms19082418
Metronidazole Causes Skeletal Muscle Atrophy and Modulates Muscle Chronometabolism
Abstract
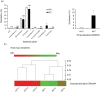
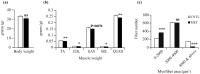
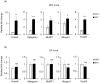
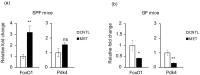
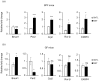
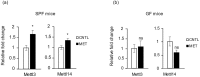
Antibiotics lead to increased susceptibility to colonization by pathogenic organisms, with different effects on the host-microbiota relationship. Here, we show that metronidazole treatment of specific pathogen-free (SPF) mice results in a significant increase of the bacterial phylum Proteobacteria in fecal pellets. Furthermore, metronidazole in SPF mice decreases hind limb muscle weight and results in smaller fibers in the tibialis anterior muscle. In the gastrocnemius muscle, metronidazole causes upregulation of Hdac4, myogenin, MuRF1, and atrogin1, which are implicated in skeletal muscle neurogenic atrophy. Metronidazole in SPF mice also upregulates skeletal muscle FoxO3, described as involved in apoptosis and muscle regeneration. Of note, alteration of the gut microbiota results in increased expression of the muscle core clock and effector genes Cry2, Ror-β, and E4BP4. PPARγ and one of its important target genes, adiponectin, are also upregulated by metronidazole. Metronidazole in germ-free (GF) mice increases the expression of other core clock genes, such as Bmal1 and Per2, as well as the metabolic regulators FoxO1 and Pdk4, suggesting a microbiota-independent pharmacologic effect. In conclusion, metronidazole in SPF mice results in skeletal muscle atrophy and changes the expression of genes involved in the muscle peripheral circadian rhythm machinery and metabolic regulation.
Keywords: circadian rhythm; gut dysbiosis; metronidazole; skeletal muscle atrophy.
Conflict of interest statement
All authors declare no conflict of interest.
Figures

Similar articles
-
Atypical expression of circadian clock genes in denervated mouse skeletal muscle.Chronobiol Int. 2015 May;32(4):486-96. doi: 10.3109/07420528.2014.1003350. Epub 2015 Mar 23. Chronobiol Int. 2015. PMID: 25798696
-
Food deprivation during active phase induces skeletal muscle atrophy via IGF-1 reduction in mice.Arch Biochem Biophys. 2019 Nov 30;677:108160. doi: 10.1016/j.abb.2019.108160. Epub 2019 Oct 19. Arch Biochem Biophys. 2019. PMID: 31639326
-
Expression of MuRF1 or MuRF2 is essential for the induction of skeletal muscle atrophy and dysfunction in a murine pulmonary hypertension model.Skelet Muscle. 2020 Apr 27;10(1):12. doi: 10.1186/s13395-020-00229-2. Skelet Muscle. 2020. PMID: 32340625 Free PMC article.
-
PPARs and Microbiota in Skeletal Muscle Health and Wasting.Int J Mol Sci. 2020 Oct 29;21(21):8056. doi: 10.3390/ijms21218056. Int J Mol Sci. 2020. PMID: 33137899 Free PMC article. Review.
-
Do not neglect the role of circadian rhythm in muscle atrophy.Ageing Res Rev. 2020 Nov;63:101155. doi: 10.1016/j.arr.2020.101155. Epub 2020 Aug 31. Ageing Res Rev. 2020. PMID: 32882420 Review.
Cited by
-
Gut-muscle axis mechanism of exercise prevention of sarcopenia.Front Nutr. 2024 Aug 16;11:1418778. doi: 10.3389/fnut.2024.1418778. eCollection 2024. Front Nutr. 2024. PMID: 39221163 Free PMC article. Review.
-
Pathogenesis of Sarcopenia in Chronic Kidney Disease-The Role of Inflammation, Metabolic Dysregulation, Gut Dysbiosis, and microRNA.Int J Mol Sci. 2024 Aug 3;25(15):8474. doi: 10.3390/ijms25158474. Int J Mol Sci. 2024. PMID: 39126043 Free PMC article. Review.
-
Impact of a Whole-Food, High-Soluble Fiber Diet on the Gut-Muscle Axis in Aged Mice.Nutrients. 2024 Apr 28;16(9):1323. doi: 10.3390/nu16091323. Nutrients. 2024. PMID: 38732569 Free PMC article.
-
Bacteroidota inhibit microglia clearance of amyloid-beta and promote plaque deposition in Alzheimer's disease mouse models.Nat Commun. 2024 May 8;15(1):3872. doi: 10.1038/s41467-024-47683-w. Nat Commun. 2024. PMID: 38719797 Free PMC article.
-
Effect of gut microbiome modulation on muscle function and cognition: the PROMOTe randomised controlled trial.Nat Commun. 2024 Feb 29;15(1):1859. doi: 10.1038/s41467-024-46116-y. Nat Commun. 2024. PMID: 38424099 Free PMC article. Clinical Trial.
References
-
- Rejinders D., Goossens G.H., Hermes G.D., Neis E.P., van der Beek C.M., Most J., Lenaerts K., Kootte R.S., Nieuwdorp M., Groen A.K., et al. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: A randomized double-blind placebo-controlled trial. Cell Metab. 2016;24:63–74. doi: 10.1016/j.cmet.2016.06.016. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

