Identification of target genes in cardiomyopathy with fibrosis and cardiac remodeling
- PMID: 30115125
- PMCID: PMC6094872
- DOI: 10.1186/s12929-018-0459-8
Identification of target genes in cardiomyopathy with fibrosis and cardiac remodeling
Abstract
Background: Identify genes probably associated with chronic heart failure and predict potential target genes for dilated cardiomyopathy using bioinformatics analyses.
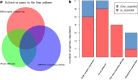
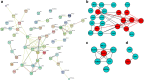
Methods: Gene expression profiles (series number GSE3585 and GSE42955) of cardiomyopathy patients and healthy controls were downloaded from the Expression Omnibus Gene (GEO) database. Differential expression of genes (DEGS) between the two groups of total 14 cardiomyopathy patients and 10 healthy controls were subsequently identified by limma package of R. Database for Annotation, Visualization, and Integrated Discovery (DAVID Tool), which is an analysis of enriched biological processes. Search Tool for the Retrieval Interacting Genes (STRING) was used as well for the analysis of protein-protein interaction network (PPI). Prediction of the potential drugs was suggested based on the preliminarily identified genes using Connectivity Map (CMap).
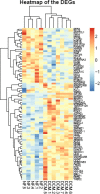
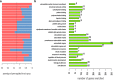
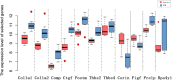
Results: Eighty-nine DEGs were identified (57 up-regulated and 32 down-regulated). The most enrichment Gene Ontology (GO) terms (P < 0.05) contain genes involved in extracellular matrix (ECM) and biological adhesion signal pathways (P < 0.05, ES > 1.5) such as ECM-receptors, focal adhesion and transforming growth factor beta (TGF-β), etc. Fifty-one differentially expressed genes were found to encode interacting proteins. Eleven key genes along with related transcription factors were identified including CTGF, POSTN, CORIN, FIGF, etc. CONCLUSION: Bioinformatics-based analyses reveal the targeted genes probably associated with cardiomyopathy, which provide clues for pharmacological therapies aiming at the targets.
Keywords: Bioinformatics; Dilated cardiomyopathy; Heart failure; Microarray.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Integrated bioinformatics analysis reveals novel key biomarkers and potential candidate small molecule drugs in gastric cancer.Pathol Res Pract. 2019 May;215(5):1038-1048. doi: 10.1016/j.prp.2019.02.012. Epub 2019 Feb 28. Pathol Res Pract. 2019. PMID: 30975489
-
Analyzing gene expression profiles in dilated cardiomyopathy via bioinformatics methods.Braz J Med Biol Res. 2016 Oct 10;49(10):e4897. doi: 10.1590/1414-431X20164897. Braz J Med Biol Res. 2016. PMID: 27737314 Free PMC article.
-
Identification of the molecular mechanisms underlying dilated cardiomyopathy via bioinformatic analysis of gene expression profiles.Exp Ther Med. 2017 Jan;13(1):273-279. doi: 10.3892/etm.2016.3953. Epub 2016 Dec 5. Exp Ther Med. 2017. PMID: 28123501 Free PMC article.
-
Identification of key genes and crucial modules associated with coronary artery disease by bioinformatics analysis.Int J Mol Med. 2014 Sep;34(3):863-9. doi: 10.3892/ijmm.2014.1817. Epub 2014 Jun 23. Int J Mol Med. 2014. PMID: 24969630
-
Identification of human prolactinoma related genes by DNA microarray.J Cancer Res Ther. 2014 Jul-Sep;10(3):544-8. doi: 10.4103/0973-1482.137962. J Cancer Res Ther. 2014. PMID: 25313736
Cited by
-
Preventive and treatment efficiency of dendrosomal nano-curcumin against ISO-induced cardiac fibrosis in mouse model.PLoS One. 2024 Oct 10;19(10):e0311817. doi: 10.1371/journal.pone.0311817. eCollection 2024. PLoS One. 2024. PMID: 39388499 Free PMC article.
-
Corin: A Key Mediator in Sodium Homeostasis, Vascular Remodeling, and Heart Failure.Biology (Basel). 2022 May 7;11(5):717. doi: 10.3390/biology11050717. Biology (Basel). 2022. PMID: 35625445 Free PMC article. Review.
-
Predicting the Key Genes Involved in Aortic Valve Calcification Through Integrated Bioinformatics Analysis.Front Genet. 2021 May 11;12:650213. doi: 10.3389/fgene.2021.650213. eCollection 2021. Front Genet. 2021. PMID: 34046056 Free PMC article.
-
Macrophage, a potential targeted therapeutic immune cell for cardiomyopathy.Front Cell Dev Biol. 2022 Sep 30;10:908790. doi: 10.3389/fcell.2022.908790. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36247005 Free PMC article. Review.
-
Serum discrimination and phenotype assessment of coronary artery disease patents with and without type 2 diabetes prior to coronary artery bypass graft surgery.PLoS One. 2020 Aug 5;15(8):e0234539. doi: 10.1371/journal.pone.0234539. eCollection 2020. PLoS One. 2020. PMID: 32756554 Free PMC article.
References
-
- Pinto YM, Elliott PM, Arbustini E, Adler Y, Anastasakis A, Böhm M, Duboc D, Gimeno J, de Groote P, Imazio M. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37(23):1850–1858. doi: 10.1093/eurheartj/ehv727. - DOI - PubMed
-
- Barth AS, Kuner R, Buness A, Ruschhaupt M, Merk S, Zwermann L, Kääb S, Kreuzer E, Steinbeck G, Mansmann U. Identification of a common gene expression signature in dilated cardiomyopathy across independent microarray studies. J Am Coll Cardiol. 2006;48:1610–1617. doi: 10.1016/j.jacc.2006.07.026. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

