A Bioconductor workflow for processing and analysing spatial proteomics data
- PMID: 30079225
- PMCID: PMC6053703
- DOI: 10.12688/f1000research.10411.2
A Bioconductor workflow for processing and analysing spatial proteomics data
Abstract
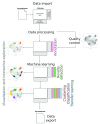
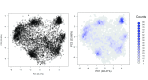
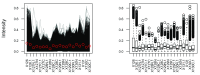
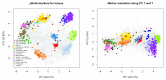
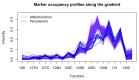
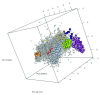
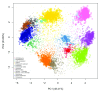
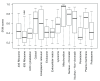
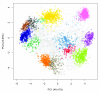
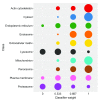
Spatial proteomics is the systematic study of protein sub-cellular localisation. In this workflow, we describe the analysis of a typical quantitative mass spectrometry-based spatial proteomics experiment using the MSnbase and pRoloc Bioconductor package suite. To walk the user through the computational pipeline, we use a recently published experiment predicting protein sub-cellular localisation in pluripotent embryonic mouse stem cells. We describe the software infrastructure at hand, importing and processing data, quality control, sub-cellular marker definition, visualisation and interactive exploration. We then demonstrate the application and interpretation of statistical learning methods, including novelty detection using semi-supervised learning, classification, clustering and transfer learning and conclude the pipeline with data export. The workflow is aimed at beginners who are familiar with proteomics in general and spatial proteomics in particular.
Keywords: Bioconductor; R Package; machine learning; mass spectromery; protein sub-cellular localisation; proteomics; spatial proteomics; transfer learning.
Conflict of interest statement
Competing interests: No competing interests were dislcosed.
Figures

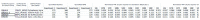




Similar articles
-
A Bioconductor workflow for the Bayesian analysis of spatial proteomics.F1000Res. 2019 Apr 11;8:446. doi: 10.12688/f1000research.18636.1. eCollection 2019. F1000Res. 2019. PMID: 31119032 Free PMC article.
-
Learning from Heterogeneous Data Sources: An Application in Spatial Proteomics.PLoS Comput Biol. 2016 May 13;12(5):e1004920. doi: 10.1371/journal.pcbi.1004920. eCollection 2016 May. PLoS Comput Biol. 2016. PMID: 27175778 Free PMC article.
-
Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata.Bioinformatics. 2014 May 1;30(9):1322-4. doi: 10.1093/bioinformatics/btu013. Epub 2014 Jan 11. Bioinformatics. 2014. PMID: 24413670 Free PMC article.
-
SubCellBarCode: integrated workflow for robust spatial proteomics by mass spectrometry.Nat Protoc. 2022 Aug;17(8):1832-1867. doi: 10.1038/s41596-022-00699-2. Epub 2022 Jun 22. Nat Protoc. 2022. PMID: 35732783 Review.
-
An end-to-end workflow for multiplexed image processing and analysis.Nat Protoc. 2023 Nov;18(11):3565-3613. doi: 10.1038/s41596-023-00881-0. Epub 2023 Oct 10. Nat Protoc. 2023. PMID: 37816904 Review.
Cited by
-
Reduced mitochondria provide an essential function for the cytosolic methionine cycle.Curr Biol. 2022 Dec 5;32(23):5057-5068.e5. doi: 10.1016/j.cub.2022.10.028. Epub 2022 Nov 7. Curr Biol. 2022. PMID: 36347252 Free PMC article.
-
Spatiotemporal proteomic profiling of the pro-inflammatory response to lipopolysaccharide in the THP-1 human leukaemia cell line.Nat Commun. 2021 Oct 1;12(1):5773. doi: 10.1038/s41467-021-26000-9. Nat Commun. 2021. PMID: 34599159 Free PMC article.
-
A Bioconductor workflow for the Bayesian analysis of spatial proteomics.F1000Res. 2019 Apr 11;8:446. doi: 10.12688/f1000research.18636.1. eCollection 2019. F1000Res. 2019. PMID: 31119032 Free PMC article.
-
Mapping diversity in African trypanosomes using high resolution spatial proteomics.Nat Commun. 2023 Jul 21;14(1):4401. doi: 10.1038/s41467-023-40125-z. Nat Commun. 2023. PMID: 37479728 Free PMC article.
-
Comparative Analysis of Quantitative Mass Spectrometric Methods for Subcellular Proteomics.J Proteome Res. 2020 Apr 3;19(4):1718-1730. doi: 10.1021/acs.jproteome.9b00862. Epub 2020 Mar 5. J Proteome Res. 2020. PMID: 32134668 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

