A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte
- PMID: 30069046
- PMCID: PMC6108322
- DOI: 10.1038/s41586-018-0394-6
A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte
Abstract
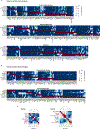
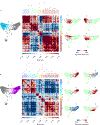
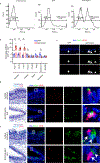
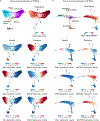
The functions of epithelial tissues are dictated by the types, abundance and distribution of the differentiated cells they contain. Attempts to restore tissue function after damage require knowledge of how physiological tasks are distributed among cell types, and how cell states vary between homeostasis, injury-repair and disease. In the conducting airway, a heterogeneous basal cell population gives rise to specialized luminal cells that perform mucociliary clearance1. Here we perform single-cell profiling of human bronchial epithelial cells and mouse tracheal epithelial cells to obtain a comprehensive census of cell types in the conducting airway and their behaviour in homeostasis and regeneration. Our analysis reveals cell states that represent known and novel cell populations, delineates their heterogeneity and identifies distinct differentiation trajectories during homeostasis and tissue repair. Finally, we identified a novel, rare cell type that we call the 'pulmonary ionocyte', which co-expresses FOXI1, multiple subunits of the vacuolar-type H+-ATPase (V-ATPase) and CFTR, the gene that is mutated in cystic fibrosis. Using immunofluorescence, modulation of signalling pathways and electrophysiology, we show that Notch signalling is necessary and FOXI1 expression is sufficient to drive the production of the pulmonary ionocyte, and that the pulmonary ionocyte is a major source of CFTR activity in the conducting airway epithelium.
Conflict of interest statement
Figures








Comment in
-
Profile of an unknown airway cell.Nature. 2018 Aug;560(7718):313-314. doi: 10.1038/d41586-018-05813-7. Nature. 2018. PMID: 30097657 No abstract available.
-
Pulmonary Ionocytes Challenge the Paradigm in Cystic Fibrosis.Trends Pharmacol Sci. 2018 Oct;39(10):852-854. doi: 10.1016/j.tips.2018.08.005. Epub 2018 Sep 10. Trends Pharmacol Sci. 2018. PMID: 30213439
Similar articles
-
Transgenic ferret models define pulmonary ionocyte diversity and function.Nature. 2023 Sep;621(7980):857-867. doi: 10.1038/s41586-023-06549-9. Epub 2023 Sep 20. Nature. 2023. PMID: 37730992 Free PMC article.
-
A revised airway epithelial hierarchy includes CFTR-expressing ionocytes.Nature. 2018 Aug;560(7718):319-324. doi: 10.1038/s41586-018-0393-7. Epub 2018 Aug 1. Nature. 2018. PMID: 30069044 Free PMC article.
-
Ionocytes and CFTR Chloride Channel Expression in Normal and Cystic Fibrosis Nasal and Bronchial Epithelial Cells.Cells. 2020 Sep 13;9(9):2090. doi: 10.3390/cells9092090. Cells. 2020. PMID: 32933106 Free PMC article.
-
Cystic Fibrosis and the Cells of the Airway Epithelium: What Are Ionocytes and What Do They Do?Annu Rev Pathol. 2022 Jan 24;17:23-46. doi: 10.1146/annurev-pathol-042420-094031. Epub 2021 Aug 26. Annu Rev Pathol. 2022. PMID: 34437820 Free PMC article. Review.
-
Using single-cell RNA sequencing to unravel cell lineage relationships in the respiratory tract.Biochem Soc Trans. 2020 Feb 28;48(1):327-336. doi: 10.1042/BST20191010. Biochem Soc Trans. 2020. PMID: 31922198 Review.
Cited by
-
Life-long functional regeneration of in vivo airway epithelium by the engraftment of airway basal stem cells.Nat Protoc. 2024 Nov 5. doi: 10.1038/s41596-024-01067-y. Online ahead of print. Nat Protoc. 2024. PMID: 39501108 Review.
-
Reinitiating lung development: a novel approach in the management of bronchopulmonary dysplasia.Respir Res. 2024 Oct 24;25(1):384. doi: 10.1186/s12931-024-02996-8. Respir Res. 2024. PMID: 39449014 Free PMC article. Review.
-
CTISL: a dynamic stacking multi-class classification approach for identifying cell types from single-cell RNA-seq data.Bioinformatics. 2024 Feb 1;40(2):btae063. doi: 10.1093/bioinformatics/btae063. Bioinformatics. 2024. PMID: 38317054 Free PMC article.
-
Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes.PLoS Biol. 2021 Mar 17;19(3):e3001143. doi: 10.1371/journal.pbio.3001143. eCollection 2021 Mar. PLoS Biol. 2021. PMID: 33730024 Free PMC article.
-
Pharmacological Improvement of Cystic Fibrosis Transmembrane Conductance Regulator Function Rescues Airway Epithelial Homeostasis and Host Defense in Children with Cystic Fibrosis.Am J Respir Crit Care Med. 2024 Jun 1;209(11):1338-1350. doi: 10.1164/rccm.202310-1836OC. Am J Respir Crit Care Med. 2024. PMID: 38259174 Free PMC article.
References
-
- Hong KU, Reynolds SD, Watkins S, Fuchs E & Stripp BR In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol 286, L643–L649 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

