The interplay between H2A.Z and H3K9 methylation in regulating HP1α binding to linker histone-containing chromatin
- PMID: 30007360
- PMCID: PMC6182156
- DOI: 10.1093/nar/gky632
The interplay between H2A.Z and H3K9 methylation in regulating HP1α binding to linker histone-containing chromatin
Abstract
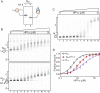
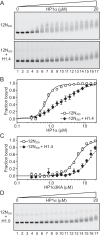
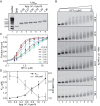
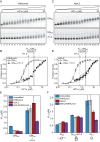
One of the most intensively studied chromatin binding factors is HP1α. HP1α is associated with silenced, heterochromatic regions of the genome and binds to H3K9me3. While H3K9me3 is necessary for HP1α recruitment to heterochromatin, it is becoming apparent that it is not sufficient suggesting that additional factors are involved. One candidate proposed as a potential regulator of HP1α recruitment is the linker histone H1.4. Changes to the underlying make-up of chromatin, such as the incorporation of the histone variant H2A.Z, has also been linked with regulating HP1 binding to chromatin. Here, we rigorously dissected the effects of H1.4, H2A.Z and H3K9me3 on the nucleosome binding activity of HP1α in vitro employing arrays, mononucleosomes and nucleosome core particles. Unexpectedly, histone H1.4 impedes the binding of HP1α but strikingly, this inhibition is partially relieved by the incorporation of both H2A.Z and H3K9me3 but only in the context of arrays or nucleosome core particles. Our data suggests that there are two modes of interaction of HP1α with nucleosomes. The first primary mode is through interactions with linker DNA. However, when linker DNA is missing or occluded by linker histones, HP1α directly interacts with the nucleosome core and this interaction is enhanced by H2A.Z with H3K9me3.
Figures

Similar articles
-
Structural mechanism of HP1⍺-dependent transcriptional repression and chromatin compaction.Structure. 2024 Nov 7;32(11):2094-2106.e6. doi: 10.1016/j.str.2024.09.013. Epub 2024 Oct 8. Structure. 2024. PMID: 39383876 Free PMC article.
-
Structural Basis of Heterochromatin Formation by Human HP1.Mol Cell. 2018 Feb 1;69(3):385-397.e8. doi: 10.1016/j.molcel.2017.12.011. Epub 2018 Jan 11. Mol Cell. 2018. PMID: 29336876
-
Hinge and chromoshadow of HP1α participate in recognition of K9 methylated histone H3 in nucleosomes.J Mol Biol. 2013 Jan 9;425(1):54-70. doi: 10.1016/j.jmb.2012.10.018. Epub 2012 Nov 6. J Mol Biol. 2013. PMID: 23142645
-
Chromatin structures condensed by linker histones.Essays Biochem. 2019 Apr 23;63(1):75-87. doi: 10.1042/EBC20180056. Print 2019 Apr 23. Essays Biochem. 2019. PMID: 31015384 Review.
-
Histone variants--the structure behind the function.Brief Funct Genomic Proteomic. 2006 Sep;5(3):228-43. doi: 10.1093/bfgp/ell020. Epub 2006 May 10. Brief Funct Genomic Proteomic. 2006. PMID: 16772274 Review.
Cited by
-
Depletion of HP1α alters the mechanical properties of MCF7 nuclei.Biophys J. 2021 Jul 6;120(13):2631-2643. doi: 10.1016/j.bpj.2021.05.017. Epub 2021 Jun 2. Biophys J. 2021. PMID: 34087208 Free PMC article.
-
Coordinated changes in gene expression, H1 variant distribution and genome 3D conformation in response to H1 depletion.Nucleic Acids Res. 2022 Apr 22;50(7):3892-3910. doi: 10.1093/nar/gkac226. Nucleic Acids Res. 2022. PMID: 35380694 Free PMC article.
-
Chromatin remodeler Dmp18 regulates apoptosis by controlling H2Av incorporation in Drosophila imaginal disc development.PLoS Genet. 2022 Sep 27;18(9):e1010395. doi: 10.1371/journal.pgen.1010395. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36166470 Free PMC article.
-
DNA sequence-dependent formation of heterochromatin nanodomains.Nat Commun. 2022 Apr 6;13(1):1861. doi: 10.1038/s41467-022-29360-y. Nat Commun. 2022. PMID: 35387992 Free PMC article.
-
How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance.Cells. 2020 Jun 12;9(6):1460. doi: 10.3390/cells9061460. Cells. 2020. PMID: 32545538 Free PMC article. Review.
References
-
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012; 148:458–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

